Functions/Subroutines | |
| subroutine | initialize_assim_categories_hype (assimInfo, stateinfo) |
| subroutine | get_statetype_from_category (assimInfo, stateinfo, catname, svname, stname) |
| subroutine | set_assimilate (assimInfo, stateinfo, ivar, astatus) |
| subroutine | get_assimilate_for_category (assimInfo, catname, astatus) |
| subroutine | checkmodelstructure_states_hype2 (nx, stateinfo) |
| subroutine | get_various_hypestate_assimilation_parameters (assimInfo, stname, svname, limits, coordID, locID, transformation, lambda, epsilon) |
| subroutine | allocate_and_initialize_model_state_ensembles_hype2 (ne, assimVar, varID, assimInfo, stateinfo) |
| subroutine | calculate_coordinates_for_upstream_area (n, x, y, z) |
| subroutine | calculate_coordinates_for_aquifer (ia, x, y) |
| subroutine | allocate_and_initialize_model_forcing_ensembles_hype (ne, assimVar, varID, assimInfo, nInp, ITYPE, IID, IMIN, IMAX, ISIGMA, IMINSIGMA, ISEMIM, IRESTM, ILSCALE, IGRIDSIZE, ICORRTYPE, XC, YC, ITAU) |
| subroutine | get_various_hypeoutvar_assimilation_parameters (assimInfo, outid, outMin, outMax, coordID, locID, transformation, lambda, epsilon) |
| subroutine | allocate_and_initialize_model_auxiliary_ensembles_hype (ne, assimVar, varID, assimInfo, outid, Coordinates) |
| subroutine | allocate_and_initialize_observation_ensembles_hype (ne, assimVar, varID, obsid, OIDD, OIDDOV, OIDHXOV, OTYPE, OMIN, OMAX, OSIGMA, OMINSIGMA, OSEMIM, ORESTM, OLSCALE, OGRIDSIZE, OCORRTYPE, Coordinates, OCOORDID, OTRANSFORMATION, OEPSILON, missing) |
| subroutine | assim_initialize (fdir, dir, rdir, conductinibin, assimData, ne, stateinfo, noutvar, n_Result) |
| subroutine | model_to_ensemble2 (i_ens, assimX, assimA, nA, stateinfo, skipX) |
| subroutine | writeriverq_to_x (xriverq, ni, nj, nl, k, i_ens, assimVar) |
| subroutine | writecriver_to_x (xcriver, nh, ni, nj, nl, k, i_ens, assimVar) |
| subroutine | write1d_to_x (x1d, ni, k, i_ens, assimVar) |
| subroutine | write2d_to_x (x2d, ni, nj, k, i_ens, assimVar) |
| subroutine | write3d_to_x (x3d, ni, nj, nl, k, i_ens, assimVar) |
| subroutine | write4d_to_x (x4d, ni, nj, nl, nm, k, i_ens, assimVar) |
| subroutine | ensemble_to_model2 (i_ens, nA, nF, assimX, assimA, assimF, stateinfo) |
| subroutine | writef_to_1d (x1d, ni, k, i_ens, assimVar) |
| subroutine | writex_to_1d (x1d, ni, k, i_ens, assimVar) |
| subroutine | writex_to_2d (x2d, ni, nj, k, i_ens, assimVar) |
| subroutine | writex_to_3d (x3d, ni, nj, nl, k, i_ens, assimVar) |
| subroutine | writex_to_4d (x4d, ni, nj, nl, nm, k, i_ens, assimVar) |
| subroutine | writex_to_2d_real2int (x2d, ni, nj, k, i_ens, assimVar) |
| subroutine | writex_to_riverq (xriverq, ni, nj, nl, k, i_ens, assimVar) |
| subroutine | writex_to_criver (xcriver, nh, ni, nj, nl, k, i_ens, assimVar) |
| subroutine | statistics_to_modeloutput (nA, assimA, istat, meanORmedian) |
| subroutine | meanormedian_to_model2 (nA, assimX, assimA, meanORmedian, stateinfo) |
| subroutine | writexmedian_to_1d (x1d, ni, k, assimVar, meanORmedian) |
| subroutine | writexmedian_to_2d (x2d, ni, nj, k, assimVar, meanORmedian) |
| subroutine | writexmedian_to_2d_real2int (x2d, ni, nj, k, assimVar, meanORmedian) |
| subroutine | writexmedian_to_3d (x3d, ni, nj, nl, k, assimVar, meanORmedian) |
| subroutine | writexmedian_to_4d (x4d, ni, nj, nl, nm, k, assimVar, meanORmedian) |
| subroutine | writexmedian_to_riverq (xriverq, ni, nj, nl, k, assimVar, meanORmedian) |
| subroutine | writexmedian_to_criver (xcriver, nh, ni, nj, nl, k, assimVar, meanORmedian) |
| subroutine | generate_observation_ensemble (assimData) |
| subroutine | modelobservations_to_ensemble (i_ens, assimData) |
| subroutine | generate_forcing_ensemble (assimData) |
| subroutine | finalize_assimilation (assimData) |
| subroutine | finalize_ensemble_states_outstate (dir, ns, cdate, assimData) |
Detailed Description
Module with HYPE/HYSS specific subroutines for interface between HYPE/HYSS and the EnKF Data Assimilation routines.
Function/Subroutine Documentation
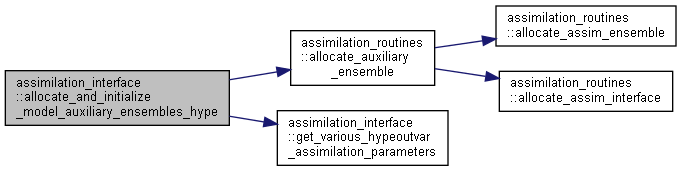
◆ allocate_and_initialize_model_auxiliary_ensembles_hype()
| subroutine assimilation_interface::allocate_and_initialize_model_auxiliary_ensembles_hype | ( | integer | ne, |
| type(assim_state_ensemble_type), dimension(:) | assimVar, | ||
| integer | varID, | ||
| type(assim_info_type), intent(in) | assimInfo, | ||
| integer | outid, | ||
| type(assim_coordinate_type), dimension(:) | Coordinates | ||
| ) |
Allocate and initialize ensembles for HYPE model auxiliary variables, i.e. outvar.
- Parameters
-
ne ensemble size varid ensemble ID outid number of auxiliaries assimvar ensemble data [in] assiminfo information, settings, etc for assimilation module
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
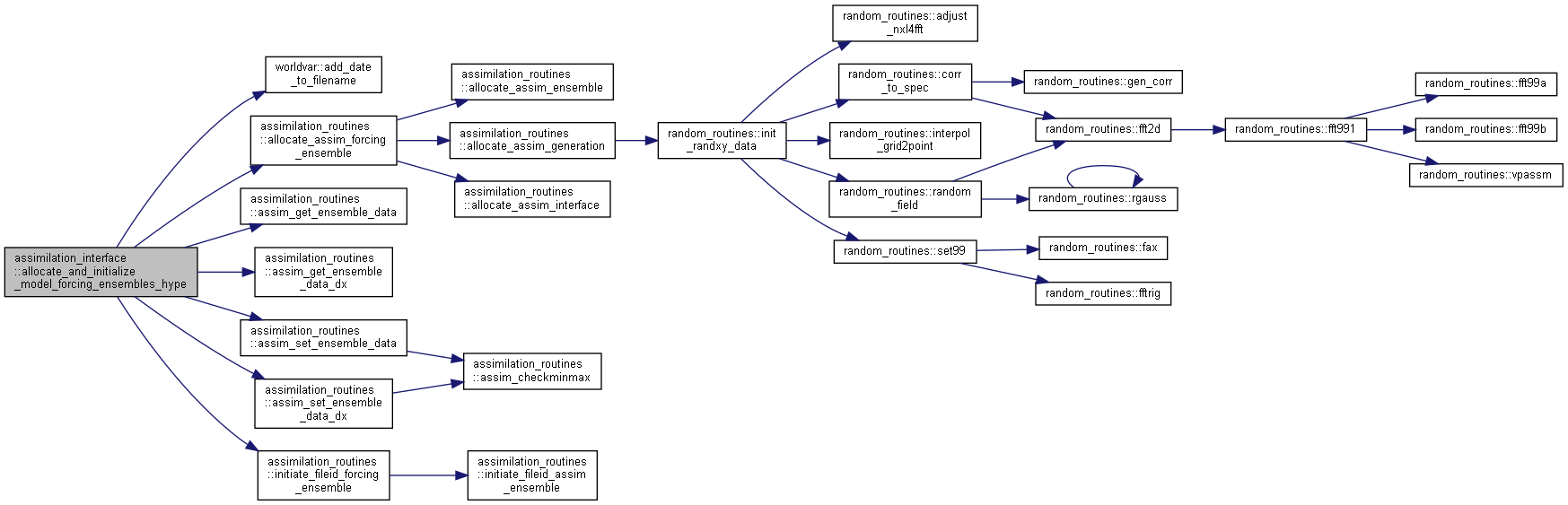
Here is the caller graph for this function:◆ allocate_and_initialize_model_forcing_ensembles_hype()
| subroutine assimilation_interface::allocate_and_initialize_model_forcing_ensembles_hype | ( | integer | ne, |
| type(assim_input_ensemble_type), dimension(:) | assimVar, | ||
| integer | varID, | ||
| type(assim_info_type), intent(in) | assimInfo, | ||
| integer | nInp, | ||
| integer, dimension(:) | ITYPE, | ||
| integer, dimension(:) | IID, | ||
| real, dimension(:) | IMIN, | ||
| real, dimension(:) | IMAX, | ||
| real, dimension(:) | ISIGMA, | ||
| real, dimension(:) | IMINSIGMA, | ||
| real, dimension(:) | ISEMIM, | ||
| real, dimension(:) | IRESTM, | ||
| real, dimension(:) | ILSCALE, | ||
| real, dimension(:) | IGRIDSIZE, | ||
| integer, dimension(:) | ICORRTYPE, | ||
| real, dimension(:) | XC, | ||
| real, dimension(:) | YC, | ||
| real, dimension(:) | ITAU | ||
| ) |
Allocate and initialize ensembles for HYPE model forcing.
- Parameters
-
ne ensemble size varid ensemble ID ninp number of forcing variables assimvar ensemble data vector [in] assiminfo information, settings, etc for assimilation module itype ensemble generation type iid variable ID icorrtype correlation function for 2D random fields
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ allocate_and_initialize_model_state_ensembles_hype2()
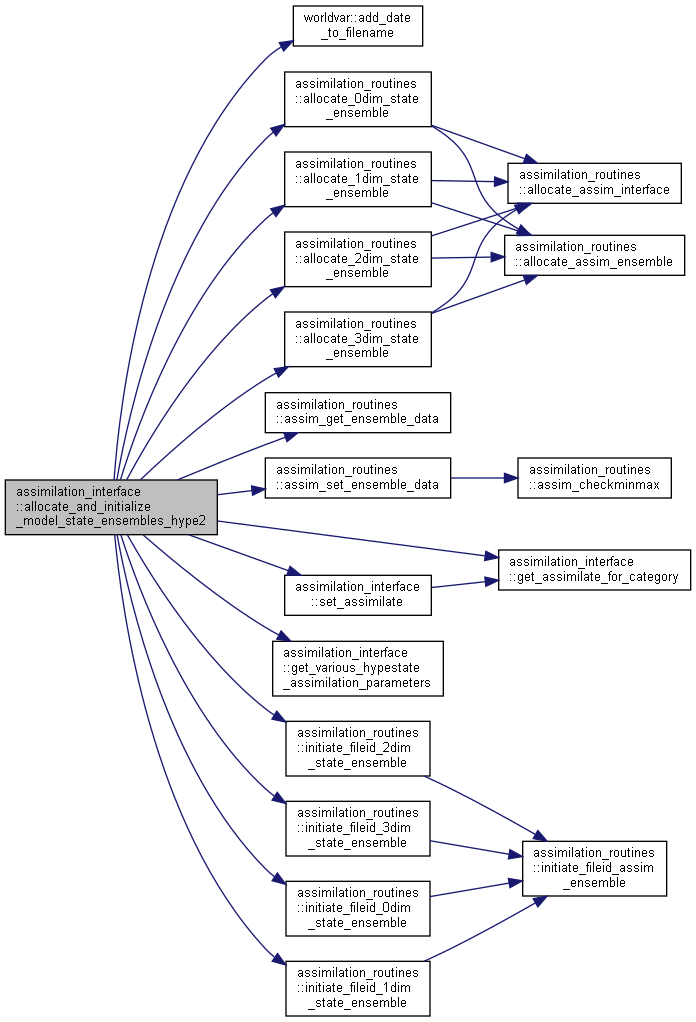
| subroutine assimilation_interface::allocate_and_initialize_model_state_ensembles_hype2 | ( | integer, intent(in) | ne, |
| type(assim_state_ensemble_type), dimension(:) | assimVar, | ||
| integer, intent(inout) | varID, | ||
| type(assim_info_type), intent(in) | assimInfo, | ||
| type(stateinfotype), dimension(:), intent(in) | stateinfo | ||
| ) |
Allocate and initialize ensembles for HYPE model states.
- Parameters
-
[in] ne ensemble size assimvar vector of state ensemble data [in,out] varid Ensemble variable ID (from 1 to number of state ensembles) [in] assiminfo information, settings, etc for assimilation module [in] stateinfo Information about state variables
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ allocate_and_initialize_observation_ensembles_hype()
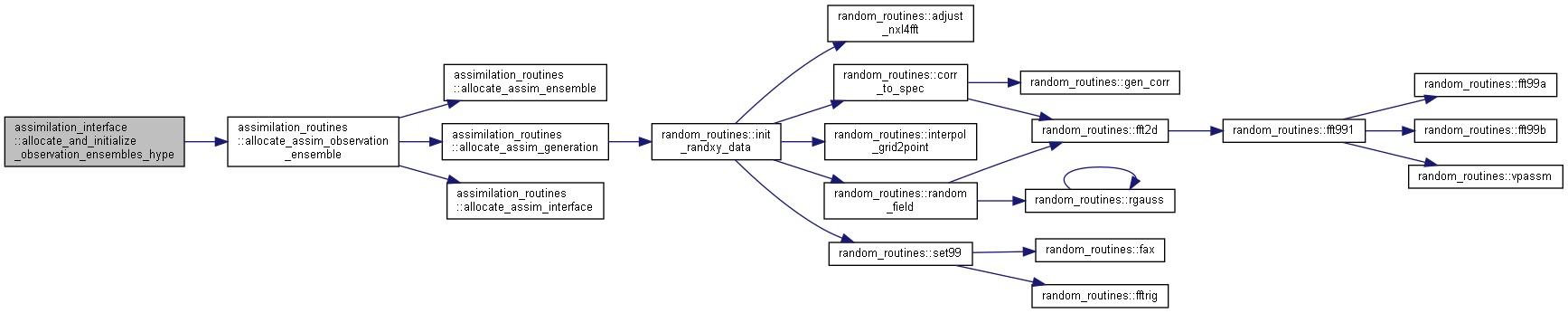
| subroutine assimilation_interface::allocate_and_initialize_observation_ensembles_hype | ( | integer | ne, |
| type(assim_input_ensemble_type), dimension(:) | assimVar, | ||
| integer | varID, | ||
| integer | obsid, | ||
| integer, dimension(:) | OIDD, | ||
| integer, dimension(:) | OIDDOV, | ||
| integer, dimension(:) | OIDHXOV, | ||
| integer, dimension(:) | OTYPE, | ||
| real, dimension(:) | OMIN, | ||
| real, dimension(:) | OMAX, | ||
| real, dimension(:) | OSIGMA, | ||
| real, dimension(:) | OMINSIGMA, | ||
| real, dimension(:) | OSEMIM, | ||
| real, dimension(:) | ORESTM, | ||
| real, dimension(:) | OLSCALE, | ||
| real, dimension(:) | OGRIDSIZE, | ||
| integer, dimension(:) | OCORRTYPE, | ||
| type(assim_coordinate_type), dimension(:) | Coordinates, | ||
| integer, dimension(:) | OCOORDID, | ||
| integer, dimension(:) | OTRANSFORMATION, | ||
| real, dimension(:) | OEPSILON, | ||
| real | missing | ||
| ) |
Allocate and initialize ensembles for HYPE model observations.
- Parameters
-
ne ensemble size varid ensemble ID obsid number of observations assimvar ensemble data otype ensemble generation type oidd observation id, referring to index in outvarid oiddov observation id, referring to index in outvar oidhxov model id, referring to index in outvar ocorrtype correlation function for 2d random number generation
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ assim_initialize()
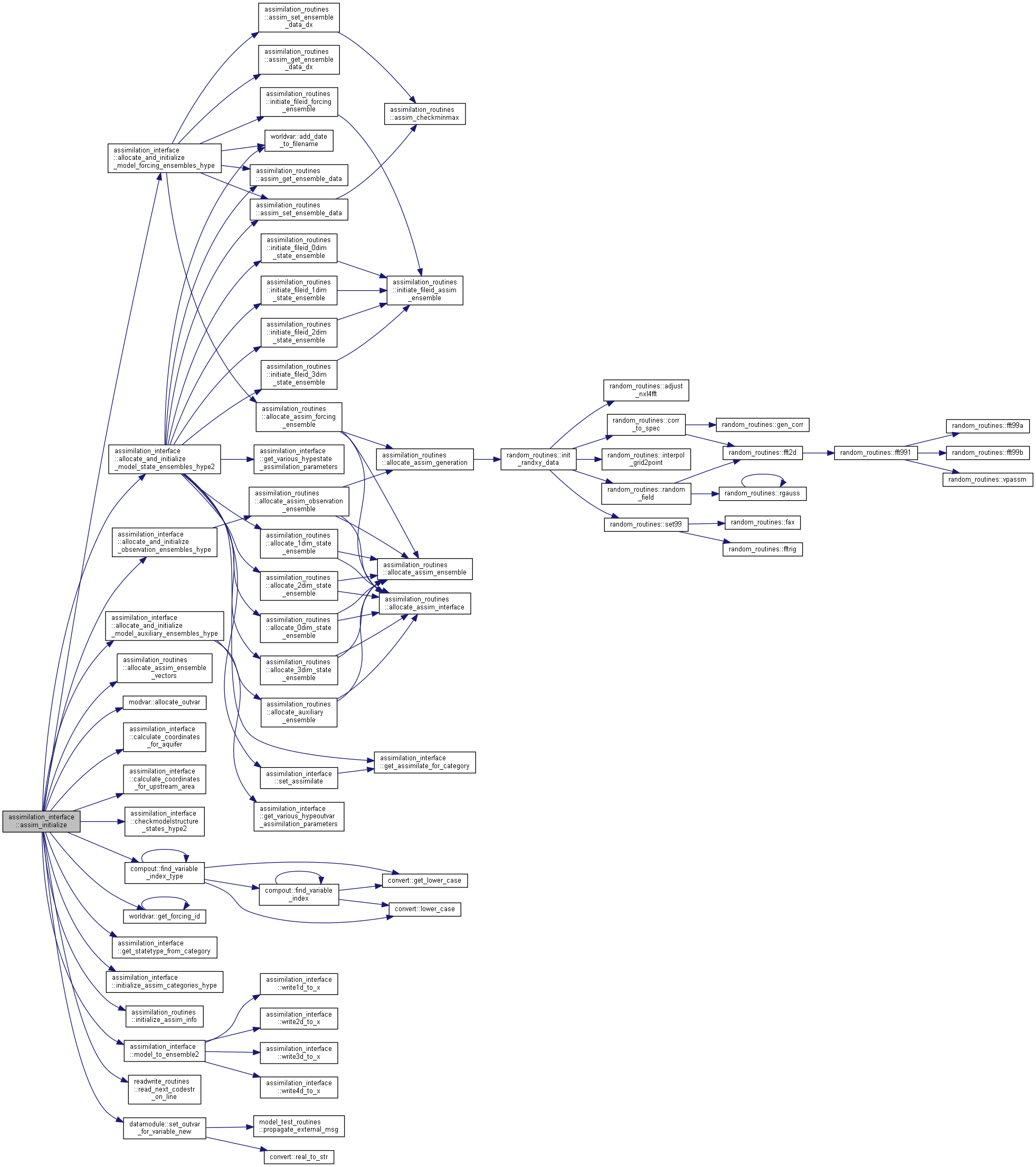
| subroutine assimilation_interface::assim_initialize | ( | character(len=maxcharpath), intent(in) | fdir, |
| character(len=maxcharpath), intent(in) | dir, | ||
| character(len=maxcharpath), intent(in) | rdir, | ||
| logical, intent(in) | conductinibin, | ||
| type(assim_data_type), intent(inout) | assimData, | ||
| integer, intent(out) | ne, | ||
| type(stateinfotype), dimension(:), intent(in) | stateinfo, | ||
| integer, intent(inout) | noutvar, | ||
| integer, intent(out) | n_Result | ||
| ) |
Initializes a assimilation simulation:
- define the basic info needed (in the code and read from input file)
- allocate the assimilation variables (model ensemble, input ensemble, etc)
- initialize assimilation variables
- Parameters
-
[in] fdir path to initial bin files with state ensemble (forcingdir) [in] dir path to AssimInfo.txt [in] rdir path to output directory (also store temporary bin files with state ensemble) [in,out] assimdata main assimilation variable containing all data [out] ne Ensemble size (read from AssimInfo.txt by this routine) [in] stateinfo Information about state variables [in,out] noutvar number of output variables to be set (so far) !CP added this [out] n_result Error status of subroutine !CP add this!
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ calculate_coordinates_for_aquifer()
| subroutine assimilation_interface::calculate_coordinates_for_aquifer | ( | integer, intent(in) | ia, |
| real, intent(out) | x, | ||
| real, intent(out) | y | ||
| ) |
Calculate (middle) coordinates for aquifers.
The coordinates are calculated as area weighted subbasin coordinates of subbasin recieving or recharging to/from the aquifer.
- Parameters
-
[in] ia index of aquifer [out] x x-coordinate of aquifer [out] y y-coordinate of aquifer
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ calculate_coordinates_for_upstream_area()
| subroutine assimilation_interface::calculate_coordinates_for_upstream_area | ( | integer, intent(in) | n, |
| real, dimension(n), intent(out) | x, | ||
| real, dimension(n), intent(out) | y, | ||
| real, dimension(n), intent(out) | z | ||
| ) |
Calculate (middle) coordinates for upstream areas.
The coordinates are calculated as area weighted subbasin coordinates of subbasin upstream of the subbasin.
- Parameters
-
[in] n index of subbasin [out] x x-coordinate of subbasin upstream area [out] y y-coordinate of subbasin upstream area [out] z z-coordinate of subbasin upstream area
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ checkmodelstructure_states_hype2()
| subroutine assimilation_interface::checkmodelstructure_states_hype2 | ( | integer, intent(out) | nx, |
| type(stateinfotype), dimension(:), intent(in) | stateinfo | ||
| ) |
A subroutine to count the number of states variables in HYPE + some other HYPEVARIABLES.
- Parameters
-
[out] nx number of state variables (each variable is size nsubbasin but aquifer states) [in] stateinfo information on HYPE state variables
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ ensemble_to_model2()
| subroutine assimilation_interface::ensemble_to_model2 | ( | integer, intent(in) | i_ens, |
| integer, intent(in) | nA, | ||
| integer, intent(in) | nF, | ||
| type(assim_state_ensemble_type), dimension(:) | assimX, | ||
| type(assim_state_ensemble_type), dimension(:) | assimA, | ||
| type(assim_input_ensemble_type), dimension(:) | assimF, | ||
| type(stateinfotype), dimension(:), intent(inout) | stateinfo | ||
| ) |
Copy all model states from the ensemble variables or files.
Purpose: Write ensemble data to the model variables (states, outvars, forcing).
Reverse of subroutine model_to_ensemble
- Parameters
-
[in] i_ens current ensemble [in] na number of auxiliaries (outvars) requested [in] nf number of forcing variables assimx ensemble data for model states assima ensemble data for auxiliaries assimf ensemble data for forcing [in,out] stateinfo Information about state variables
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ finalize_assimilation()
| subroutine assimilation_interface::finalize_assimilation | ( | type(assim_data_type), intent(in) | assimData | ) |
Closes all open binary files with ensemble state value.
- Parameters
-
[in] assimdata variable holding all nescessary ensemble data for assimilation
Algorithm
Check if bin files are being used, close them
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ finalize_ensemble_states_outstate()
| subroutine assimilation_interface::finalize_ensemble_states_outstate | ( | character(len=*), intent(in) | dir, |
| integer, intent(in) | ns, | ||
| type(datetype), intent(in) | cdate, | ||
| type(assim_data_type), intent(inout) | assimData | ||
| ) |
Saves ensemble state values for later use as starting state.
- Parameters
-
[in] dir file directory for result files [in] ns number of subbasins [in] cdate current date [in,out] assimdata variable holding all nescessary ensemble data for assimilation (will not change but possible in used subroutine)
Algorithm
Check if we should save bin files at this time
Preparations, date for file name
Ensemble HYPE state files to be saved
- Read temporary state ensemble file to memory.
- Write the data back to dated bin files in the results directory.
- Reopen temporary files for continued calculations
Ensemble forcing state files to be saved
- Read temporary forcing ensemble file to memory.
- Write the data back to dated bin files in the results directory.
- Reopen temporary files for continued calculations
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ generate_forcing_ensemble()
| subroutine assimilation_interface::generate_forcing_ensemble | ( | type(assim_data_type) | assimData | ) |
Routine to generate forcing data ensembles.
- Parameters
-
assimdata main assimilation variable containing all data
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ generate_observation_ensemble()
| subroutine assimilation_interface::generate_observation_ensemble | ( | type(assim_data_type) | assimData | ) |
Generate observation ensemble and calculate its localization.
Purpose: To check if there are available observations, and if so:
- generate the observation data ensembles
- calculate and populate corresponding localization matrices
The localization is based on x,y, and z coordinates of the subbasins where the observation is located - however, for discharge data we should actually take into account the entire upstream area somehow.
- Parameters
-
assimdata main assimilation variable containing all data
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ get_assimilate_for_category()
| subroutine assimilation_interface::get_assimilate_for_category | ( | type(assim_info_type), intent(in) | assimInfo, |
| character(len=*), intent(in) | catname, | ||
| logical, intent(out) | astatus | ||
| ) |
Find out if the given category is to be assimilated.
- Parameters
-
[in] assiminfo information on assimilation simulation [in] catname category name [out] astatus flag if assimilate this category
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ get_statetype_from_category()
| subroutine assimilation_interface::get_statetype_from_category | ( | type(assim_info_type), intent(in) | assimInfo, |
| type(stateinfotype), dimension(:), intent(in) | stateinfo, | ||
| character(len=*), intent(in) | catname, | ||
| character(len=*), intent(in) | svname, | ||
| character(len=*), intent(out) | stname | ||
| ) |
Get name of state type for a given state variable in a category.
- Parameters
-
[in] assiminfo information on assimilation simulation [in] stateinfo information on HYPE state variables [in] catname category name [in] svname state variable name [out] stname state type name
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ get_various_hypeoutvar_assimilation_parameters()
| subroutine assimilation_interface::get_various_hypeoutvar_assimilation_parameters | ( | type(assim_info_type), intent(in) | assimInfo, |
| integer, intent(in) | outid, | ||
| real, intent(out) | outMin, | ||
| real, intent(out) | outMax, | ||
| integer, intent(inout) | coordID, | ||
| integer, intent(inout) | locID, | ||
| integer, intent(inout) | transformation, | ||
| real, intent(inout) | lambda, | ||
| real, intent(inout) | epsilon | ||
| ) |
Function to get various assimilation parameters for HYPE outvar variables.
- Parameters
-
[in] assiminfo information, settings, etc for assimilation module [in] outid rownr in outvar table [in,out] coordid ID for coordinate system for this [in,out] locid ID for localization matrix for this [out] outmin minimum value for this [out] outmax maximum value for this [in,out] transformation transformation option (0,1,2,3 ~ none, log, yeo-johnson,logit [in,out] lambda minimum value for this [in,out] epsilon maximum value for this
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ get_various_hypestate_assimilation_parameters()
| subroutine assimilation_interface::get_various_hypestate_assimilation_parameters | ( | type(assim_info_type), intent(in) | assimInfo, |
| character(len=20), intent(in) | stname, | ||
| character(len=20), intent(in) | svname, | ||
| real, dimension(:,:), intent(in) | limits, | ||
| integer, intent(out) | coordID, | ||
| integer, intent(out) | locID, | ||
| integer, intent(out) | transformation, | ||
| real, intent(out) | lambda, | ||
| real, intent(out) | epsilon | ||
| ) |
A subroutine to get various assimilation parameters for HYPE state variables.
- Parameters
-
[in] assiminfo information, settings, etc for assimilation module [in] stname state type name [in] svname state type name [in] limits array of state variable limits [out] coordid ID for coordinate system for this [out] locid ID for localization matrix for this [out] transformation transformation option (0,1,2,3 ~ none, log, yeo-johnson,logit [out] lambda minimum value for this [out] epsilon maximum value for this
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ initialize_assim_categories_hype()
| subroutine assimilation_interface::initialize_assim_categories_hype | ( | type(assim_info_type), intent(inout) | assimInfo, |
| type(stateinfotype), dimension(:), intent(in) | stateinfo | ||
| ) |
Defines the categories for assimilating groups of (state) variables for HYPE.
- Parameters
-
[in,out] assiminfo information on assimilation simulation [in] stateinfo information on HYPE state variables
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ meanormedian_to_model2()
| subroutine assimilation_interface::meanormedian_to_model2 | ( | integer, intent(in) | nA, |
| type(assim_state_ensemble_type), dimension(:) | assimX, | ||
| type(assim_state_ensemble_type), dimension(:) | assimA, | ||
| logical, intent(in) | meanORmedian, | ||
| type(stateinfotype), dimension(:), intent(inout) | stateinfo | ||
| ) |
Collect mean or median of ensemble to model states and auxiliary variables.
Purpose: write the mean or median of the ensemble to the model domain, for the standard model output files.
- Parameters
-
[in] na number of auxiliaries (outvars) requested assimx ensemble data for model states assima ensemble data for auxiliaries [in] meanormedian flag for getting mean (true) or median (false) [in,out] stateinfo Information about state variables
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ model_to_ensemble2()
| subroutine assimilation_interface::model_to_ensemble2 | ( | integer, intent(in) | i_ens, |
| type(assim_state_ensemble_type), dimension(:) | assimX, | ||
| type(assim_state_ensemble_type), dimension(:) | assimA, | ||
| integer, intent(in) | nA, | ||
| type(stateinfotype), dimension(:), intent(in) | stateinfo, | ||
| logical | skipX | ||
| ) |
Copy all current model states to the ensemble holding variables or files.
Purpose: Write model variables (states, parameters, etc) to the ensemble matrices. The user provide the necessary code to write data from the ensemble member i_ens to the ensemble matrix.
- Parameters
-
[in] i_ens current ensemble assimx ensemble data for model states assima ensemble data for auxiliaries [in] na number of auxiliaries (outvars) requested [in] stateinfo Information about state variables skipx logical switch to skip updating X (used when initializing from existing binfiles)
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ modelobservations_to_ensemble()
| subroutine assimilation_interface::modelobservations_to_ensemble | ( | integer | i_ens, |
| type(assim_data_type) | assimData | ||
| ) |
Model equivalents are transfered to the observation ensemble by the observation operator.
Purpose: This is the observation operator, aka the "H function", which fills the modelled observations ensemble (HX) with model data corresponding to the available observations (the D matrix).
Actions: Based on the observations in the current D matrix (EnkfD) the corresponding model variables are derived from the current outvar. So far, no additional transformations have been necessary, but it is straight forward to include whatever operator is needed in this function.
- Parameters
-
assimdata main assimilation variable containing all data i_ens current ensemble
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ set_assimilate()
| subroutine assimilation_interface::set_assimilate | ( | type(assim_info_type), intent(in) | assimInfo, |
| type(stateinfotype), dimension(:), intent(in) | stateinfo, | ||
| integer, intent(in) | ivar, | ||
| logical, intent(out) | astatus | ||
| ) |
Find out if the given state variable is to be assimilated.
- Parameters
-
[in] assiminfo information on assimilation simulation [in] stateinfo information on HYPE state variables [in] ivar index of current state variable [out] astatus flag if assimilate this variable
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ statistics_to_modeloutput()
| subroutine assimilation_interface::statistics_to_modeloutput | ( | integer, intent(in) | nA, |
| type(assim_state_ensemble_type), dimension(:), intent(in) | assimA, | ||
| integer, intent(in) | istat, | ||
| logical, intent(in), optional | meanORmedian | ||
| ) |
Collect wanted statistics of ensemble to output.
- Writes mean or median of the ensemble to outvar for printout
- Writes the min/max etc of the ensemble to outvar for printout to the "sequence" model output files
- Writes individual ensembles to outvar for printout to the "sequence" model output files
- Parameters
-
[in] na number of auxiliaries (outvars) requested [in] assima assimilation data on auxiliary variables (outvar) [in] istat choice of statistics, 2=min,3=max,4=0.025q,5=median,6=0.975q,7=1st ensemble, 8=2nd ensemble... [in] meanormedian flag for setting mean (true) or median (false)
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ write1d_to_x()
| subroutine assimilation_interface::write1d_to_x | ( | real, dimension(:), intent(in) | x1d, |
| integer, intent(in) | ni, | ||
| integer, intent(inout) | k, | ||
| integer, intent(in) | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
Save data for one ensemble to the state ensembles in allocated matrix or file.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ write2d_to_x()
| subroutine assimilation_interface::write2d_to_x | ( | real, dimension(:,:), intent(in) | x2d, |
| integer, intent(in) | ni, | ||
| integer, intent(in) | nj, | ||
| integer, intent(inout) | k, | ||
| integer, intent(in) | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
Save data for one ensemble to the state ensembles in allocated matrix or file.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ write3d_to_x()
| subroutine assimilation_interface::write3d_to_x | ( | real, dimension(:,:,:), intent(in) | x3d, |
| integer, intent(in) | ni, | ||
| integer, intent(in) | nj, | ||
| integer, intent(in) | nl, | ||
| integer, intent(inout) | k, | ||
| integer, intent(in) | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
Save data for one ensemble to the state ensembles in allocated matrix or file.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ write4d_to_x()
| subroutine assimilation_interface::write4d_to_x | ( | real, dimension(:,:,:,:), intent(in) | x4d, |
| integer, intent(in) | ni, | ||
| integer, intent(in) | nj, | ||
| integer, intent(in) | nl, | ||
| integer, intent(in) | nm, | ||
| integer, intent(inout) | k, | ||
| integer, intent(in) | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
Save data for one ensemble to the state ensembles in allocated matrix or file.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writecriver_to_x()
| subroutine assimilation_interface::writecriver_to_x | ( | real, dimension(nh,0:ni,nj,nl), intent(in) | xcriver, |
| integer | nh, | ||
| integer | ni, | ||
| integer | nj, | ||
| integer | nl, | ||
| integer | k, | ||
| integer | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
Save data for one ensemble to the state ensembles in allocated matrix or file.
◆ writef_to_1d()
| subroutine assimilation_interface::writef_to_1d | ( | real, dimension(:), intent(out) | x1d, |
| integer, intent(in) | ni, | ||
| integer, intent(inout) | k, | ||
| integer, intent(in) | i_ens, | ||
| type(assim_input_ensemble_type), intent(in) | assimVar | ||
| ) |
Get one ensemble member from the state ensembles.
The state ensembles may be in an allocated matrix or read from bin-file.
- Parameters
-
[in] ni dimension of matrix x1d
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writeriverq_to_x()
| subroutine assimilation_interface::writeriverq_to_x | ( | real, dimension(0:ni,nj,nl), intent(in) | xriverq, |
| integer | ni, | ||
| integer | nj, | ||
| integer | nl, | ||
| integer | k, | ||
| integer | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
Save data for one ensemble to the state ensembles in allocated matrix or file.
◆ writex_to_1d()
| subroutine assimilation_interface::writex_to_1d | ( | real, dimension(:), intent(out) | x1d, |
| integer, intent(in) | ni, | ||
| integer, intent(inout) | k, | ||
| integer, intent(in) | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:), intent(in) | assimVar | ||
| ) |
Get one ensemble member from the state ensembles.
The state ensembles may be in an allocated matrix or read from bin-file.
- Parameters
-
[in] ni dimension of matrix x1d
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writex_to_2d()
| subroutine assimilation_interface::writex_to_2d | ( | real, dimension(:,:), intent(out) | x2d, |
| integer, intent(in) | ni, | ||
| integer, intent(in) | nj, | ||
| integer, intent(inout) | k, | ||
| integer, intent(in) | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
Get one ensemble member from the state ensembles.
The state ensembles may be in an allocated matrix or read from bin-file.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writex_to_2d_real2int()
| subroutine assimilation_interface::writex_to_2d_real2int | ( | integer, dimension(:,:), intent(out) | x2d, |
| integer, intent(in) | ni, | ||
| integer, intent(in) | nj, | ||
| integer, intent(inout) | k, | ||
| integer, intent(in) | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
Get one ensemble member from the state ensembles.
The recieving matrix is an integer, so transformation may be used. The state ensembles may be in an allocated matrix or read from bin-file.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writex_to_3d()
| subroutine assimilation_interface::writex_to_3d | ( | real, dimension(:,:,:), intent(out) | x3d, |
| integer, intent(in) | ni, | ||
| integer, intent(in) | nj, | ||
| integer, intent(in) | nl, | ||
| integer, intent(inout) | k, | ||
| integer, intent(in) | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
Get one ensemble member from the state ensembles.
The state ensembles may be in an allocated matrix or read from bin-file.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writex_to_4d()
| subroutine assimilation_interface::writex_to_4d | ( | real, dimension(:,:,:,:), intent(out) | x4d, |
| integer, intent(in) | ni, | ||
| integer, intent(in) | nj, | ||
| integer, intent(in) | nl, | ||
| integer, intent(in) | nm, | ||
| integer, intent(inout) | k, | ||
| integer, intent(in) | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
Get one ensemble member from the state ensembles.
The state ensembles may be in an allocated matrix or read from bin-file.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writex_to_criver()
| subroutine assimilation_interface::writex_to_criver | ( | real, dimension(nh,0:ni,nj,nl), intent(out) | xcriver, |
| integer | nh, | ||
| integer | ni, | ||
| integer | nj, | ||
| integer | nl, | ||
| integer | k, | ||
| integer | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
◆ writex_to_riverq()
| subroutine assimilation_interface::writex_to_riverq | ( | real, dimension(0:ni,nj,nl), intent(out) | xriverq, |
| integer | ni, | ||
| integer | nj, | ||
| integer | nl, | ||
| integer | k, | ||
| integer | i_ens, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar | ||
| ) |
◆ writexmedian_to_1d()
| subroutine assimilation_interface::writexmedian_to_1d | ( | real, dimension(:), intent(out) | x1d, |
| integer, intent(in) | ni, | ||
| integer, intent(inout) | k, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar, | ||
| logical | meanORmedian | ||
| ) |
Get the median ensemble member or the mean of the ensemble members from the state ensembles.
- Parameters
-
assimvar ensemble data
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writexmedian_to_2d()
| subroutine assimilation_interface::writexmedian_to_2d | ( | real, dimension(:,:), intent(out) | x2d, |
| integer, intent(in) | ni, | ||
| integer, intent(in) | nj, | ||
| integer, intent(inout) | k, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar, | ||
| logical | meanORmedian | ||
| ) |
Get the median ensemble member or the mean of the ensemble members from the state ensembles.
- Parameters
-
assimvar ensemble data
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writexmedian_to_2d_real2int()
| subroutine assimilation_interface::writexmedian_to_2d_real2int | ( | integer, dimension(:,:), intent(out) | x2d, |
| integer, intent(in) | ni, | ||
| integer, intent(in) | nj, | ||
| integer, intent(inout) | k, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar, | ||
| logical | meanORmedian | ||
| ) |
- Parameters
-
assimvar ensemble data
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writexmedian_to_3d()
| subroutine assimilation_interface::writexmedian_to_3d | ( | real, dimension(:,:,:), intent(out) | x3d, |
| integer, intent(in) | ni, | ||
| integer, intent(in) | nj, | ||
| integer, intent(in) | nl, | ||
| integer, intent(inout) | k, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar, | ||
| logical | meanORmedian | ||
| ) |
Get the median ensemble member or the mean of the ensemble members from the state ensembles.
- Parameters
-
assimvar ensemble data
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writexmedian_to_4d()
| subroutine assimilation_interface::writexmedian_to_4d | ( | real, dimension(:,:,:,:), intent(out) | x4d, |
| integer, intent(in) | ni, | ||
| integer, intent(in) | nj, | ||
| integer, intent(in) | nl, | ||
| integer, intent(in) | nm, | ||
| integer, intent(inout) | k, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar, | ||
| logical | meanORmedian | ||
| ) |
Get the median ensemble member or the mean of the ensemble members from the state ensembles.
- Parameters
-
assimvar ensemble data
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ writexmedian_to_criver()
| subroutine assimilation_interface::writexmedian_to_criver | ( | real, dimension(nh,0:ni,nj,nl), intent(out) | xcriver, |
| integer | nh, | ||
| integer | ni, | ||
| integer | nj, | ||
| integer | nl, | ||
| integer | k, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar, | ||
| logical | meanORmedian | ||
| ) |
◆ writexmedian_to_riverq()
| subroutine assimilation_interface::writexmedian_to_riverq | ( | real, dimension(0:ni,nj,nl), intent(out) | xriverq, |
| integer | ni, | ||
| integer | nj, | ||
| integer | nl, | ||
| integer | k, | ||
| type(assim_state_ensemble_type), dimension(:) | assimVar, | ||
| logical | meanORmedian | ||
| ) |