Functions/Subroutines | |
| subroutine | meanormedian_to_ensemble (nx, assimX, meanORmedian) |
| subroutine | initialize_assim_info (assimInfo) |
| subroutine | allocate_assim_ensemble_vectors (assimData, fid_0) |
| subroutine | allocate_assim_ensemble (assimVar, nens, nvar, varID, fileID, bindir, useBinFile, locID, coordID, xini, mini, maxi, allocateOutput, missing, assimilate, iniFromBin, transformation, lambda, epsilon, dxadd) |
| subroutine | initiate_fileid_assim_ensemble (assimVar, nens, varID, fileID, useBinFile, dxadd) |
| subroutine | allocate_assim_interface (assimVar, varName, varID, modID, nSubDim, subDimID) |
| subroutine | allocate_assim_generation (assimVar, nvar, ensgen, fixsigma, semimeta, restmeta, minsigma, lscale, gridsize, corrtype, xcoord, ycoord, tau) |
| subroutine | allocate_auxiliary_ensemble (assimVar, nens, nvar, varName, xini, varID, recID, locID, coordID, fileID, bindir, useBinFile, minimum, maximum, assimilate, missing, transformation, lambda, epsilon) |
| subroutine | allocate_0dim_state_ensemble (assimVar, nens, nvar, varName, xini, varID, locID, coordID, fileID, bindir, useBinFile, narray, limitsarray, assimilate, iniFromBin, missing, transformation, lambda, epsilon) |
| subroutine | allocate_1dim_state_ensemble (assimVar, nens, nvar, varName, xini, varID, locID, coordID, fileID, bindir, useBinFile, narray, limitsarray, n1, assimilate, iniFromBin, missing, transformation, lambda, epsilon) |
| subroutine | allocate_2dim_state_ensemble (assimVar, nens, nvar, varName, xini, varID, locID, coordID, fileID, bindir, useBinfile, narray, limitsarray, n1, n2, assimilate, iniFromBin, missing, transformation, lambda, epsilon) |
| subroutine | allocate_3dim_state_ensemble (assimVar, nens, nvar, varName, xini, varID, locID, coordID, fileID, bindir, useBinFile, narray, limitsarray, n1, n2, n3, assimilate, iniFromBin, missing, transformation, lambda, epsilon) |
| subroutine | initiate_fileid_0dim_state_ensemble (assimVar, nens, varID, fileID, useBinFile) |
| subroutine | initiate_fileid_1dim_state_ensemble (assimVar, nens, varID, fileID, useBinFile, n1) |
| subroutine | initiate_fileid_2dim_state_ensemble (assimVar, nens, varID, fileID, useBinfile, n1, n2) |
| subroutine | initiate_fileid_3dim_state_ensemble (assimVar, nens, varID, fileID, useBinFile, n1, n2, n3) |
| subroutine | initiate_fileid_forcing_ensemble (assimVar, nens, varID, fileID, useBinFile, usedx) |
| subroutine | allocate_assim_forcing_ensemble (assimVar, nens, nvar, varName, xini, varID, locID, coordID, fileID, minimum, maximum, ensgen, sigma, semimeta, restmeta, minsigma, lscale, gridsize, corrtype, xcoord, ycoord, bindir, useBinFile, missing, iniFromBin, transformation, lambda, epsilon, tau, usedx) |
| subroutine | allocate_assim_observation_ensemble (assimVar, nens, nvar, varName, xini, obsID, modID, coordID, fileID, minimum, maximum, ensgen, sigma, semimeta, restmeta, minsigma, lscale, gridsize, corrtype, xcoord, ycoord, missing, transformation, lambda, epsilon) |
| subroutine | assim_checkminmax (nx, ne, ensemble, minval, maxval, missing) |
| subroutine | generate_input_ensemble (n, assimVar, missing, usedx) |
| subroutine | get_spatially_correlated_random_data2 (n, nens, assimG, X, usedx) |
| subroutine | get_random_vector_gaussian (n, a, sigma, r) |
| subroutine | matrixmatrixmultiply (mat1, mat2, matout) |
| subroutine | choldc (a, p, n, failure) |
| subroutine | cholsl (a, p, b, x, n) |
| subroutine | choleskysolution (ny, ne, P, Y, M, failure) |
| subroutine | enkf_analysis_prepare (N, ND, D, HX, R, locCyy, M, Y, HA, status) |
| subroutine | enkf_analysis_apply (N, NX, ND, M, HA, locCxy, X) |
| subroutine | assim_ensemble_statistics (xin, DIM, NN, xmean, xmins, xmaxs, xsigma, xmedian, domedian, missing) |
| subroutine | assim_get_ensemble_data (NX, NE, ensData, x) |
| subroutine | assim_set_ensemble_data (NX, NE, ensData, x, checkMinMax, missing) |
| subroutine | assim_get_ensemble_data_dx (NX, NE, ensData, dx) |
| subroutine | assim_set_ensemble_data_dx (NX, NE, ensData, x, dx, checkMinMax, missing) |
| subroutine | updateensemblestatistics (assimData, total_time) |
| subroutine | assim_forward_transform (nx, ne, x, ensdata, missing) |
| subroutine | assim_backward_transform (nx, ne, x, ensdata, missing) |
| subroutine | enkf_analysis_main (assimData) |
Detailed Description
Generic subroutines and functions for (Ensemble Kalman Filter) Data Assimilation. These functions are supposed to be general, and should not be changed Functions special for a specific model application are in assimilation_interface.f90.
Function/Subroutine Documentation
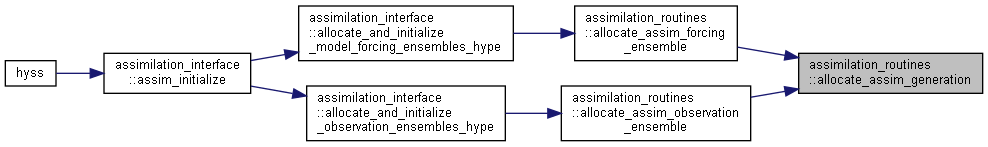
◆ allocate_0dim_state_ensemble()
| subroutine assimilation_routines::allocate_0dim_state_ensemble | ( | type(assim_state_ensemble_type) | assimVar, |
| integer | nens, | ||
| integer | nvar, | ||
| character(len=*) | varName, | ||
| real, dimension(nvar) | xini, | ||
| integer | varID, | ||
| integer | locID, | ||
| integer | coordID, | ||
| integer | fileID, | ||
| character(len=*), intent(in) | bindir, | ||
| integer, intent(in) | useBinFile, | ||
| integer, intent(in) | narray, | ||
| real, dimension(narray,2), intent(in) | limitsarray, | ||
| logical, intent(in) | assimilate, | ||
| logical, intent(in) | iniFromBin, | ||
| real | missing, | ||
| integer, intent(in) | transformation, | ||
| real, intent(in) | lambda, | ||
| real, intent(in) | epsilon | ||
| ) |
Allocate and initialize state ensemble variables; 0-dimensional.
- Parameters
-
[in] bindir path to bin-file [in] usebinfile flag for using bin-file [in] assimilate flag for including variable in assimilation [in] inifrombin flag for initializing ensemble from bin-files
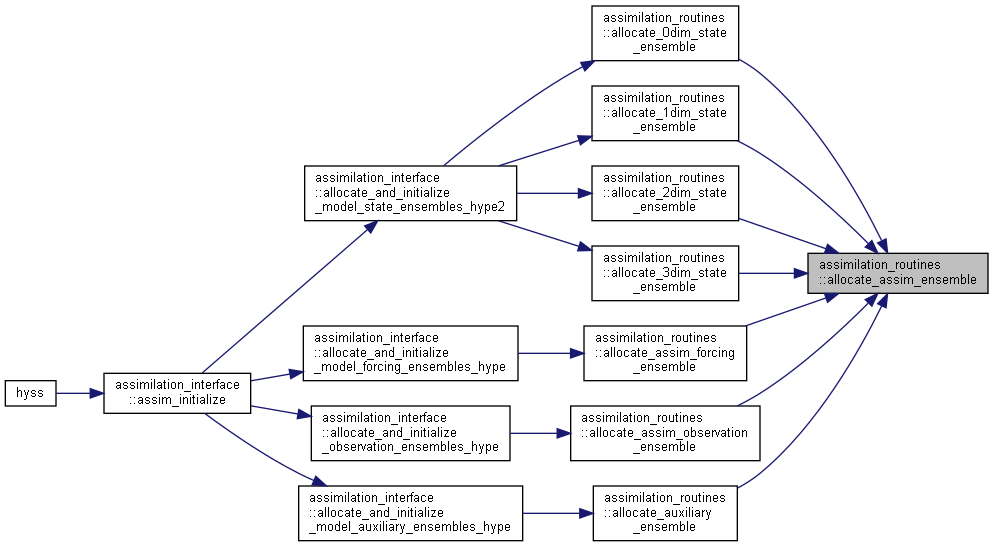
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ allocate_1dim_state_ensemble()
| subroutine assimilation_routines::allocate_1dim_state_ensemble | ( | type(assim_state_ensemble_type), dimension(:) | assimVar, |
| integer | nens, | ||
| integer | nvar, | ||
| character(len=*) | varName, | ||
| real, dimension(n1,nvar) | xini, | ||
| integer | varID, | ||
| integer | locID, | ||
| integer | coordID, | ||
| integer, dimension(:) | fileID, | ||
| character(len=*), intent(in) | bindir, | ||
| integer, intent(in) | useBinFile, | ||
| integer, intent(in) | narray, | ||
| real, dimension(narray,2), intent(in) | limitsarray, | ||
| integer | n1, | ||
| logical, intent(in) | assimilate, | ||
| logical, intent(in) | iniFromBin, | ||
| real | missing, | ||
| integer, intent(in) | transformation, | ||
| real, intent(in) | lambda, | ||
| real, intent(in) | epsilon | ||
| ) |
Allocate and initialize state ensemble variables; 1-dimensional.
- Parameters
-
[in] bindir path to bin-file [in] usebinfile flag for using bin-file [in] assimilate flag for including variable in assimilation [in] inifrombin flag for initializing ensemble from bin-files
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ allocate_2dim_state_ensemble()
| subroutine assimilation_routines::allocate_2dim_state_ensemble | ( | type(assim_state_ensemble_type), dimension(:) | assimVar, |
| integer | nens, | ||
| integer | nvar, | ||
| character(len=*) | varName, | ||
| real, dimension(n2,n1,nvar) | xini, | ||
| integer | varID, | ||
| integer | locID, | ||
| integer | coordID, | ||
| integer, dimension(:) | fileID, | ||
| character(len=*), intent(in) | bindir, | ||
| integer, intent(in) | useBinfile, | ||
| integer, intent(in) | narray, | ||
| real, dimension(narray,2), intent(in) | limitsarray, | ||
| integer | n1, | ||
| integer | n2, | ||
| logical, intent(in) | assimilate, | ||
| logical, intent(in) | iniFromBin, | ||
| real | missing, | ||
| integer, intent(in) | transformation, | ||
| real, intent(in) | lambda, | ||
| real, intent(in) | epsilon | ||
| ) |
Allocate and initialize state ensemble variables; 2-dimensional.
- Parameters
-
[in] bindir path to bin-file [in] usebinfile flag for using bin-file [in] assimilate flag for including variable in assimilation [in] inifrombin flag for initializing ensemble from bin-files
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ allocate_3dim_state_ensemble()
| subroutine assimilation_routines::allocate_3dim_state_ensemble | ( | type(assim_state_ensemble_type), dimension(:) | assimVar, |
| integer | nens, | ||
| integer | nvar, | ||
| character(len=*) | varName, | ||
| real, dimension(n3,n2,n1,nvar) | xini, | ||
| integer | varID, | ||
| integer | locID, | ||
| integer | coordID, | ||
| integer, dimension(:) | fileID, | ||
| character(len=*), intent(in) | bindir, | ||
| integer, intent(in) | useBinFile, | ||
| integer, intent(in) | narray, | ||
| real, dimension(narray,2), intent(in) | limitsarray, | ||
| integer | n1, | ||
| integer | n2, | ||
| integer | n3, | ||
| logical, intent(in) | assimilate, | ||
| logical, intent(in) | iniFromBin, | ||
| real | missing, | ||
| integer, intent(in) | transformation, | ||
| real, intent(in) | lambda, | ||
| real, intent(in) | epsilon | ||
| ) |
Allocate and initialize state ensemble variables; 3-dimensional.
- Parameters
-
[in] bindir path to bin-file [in] usebinfile flag for using bin-files [in] narray size of array [in] assimilate flag for including variable in assimilation [in] inifrombin flag for initializing ensemble from bin-files
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ allocate_assim_ensemble()
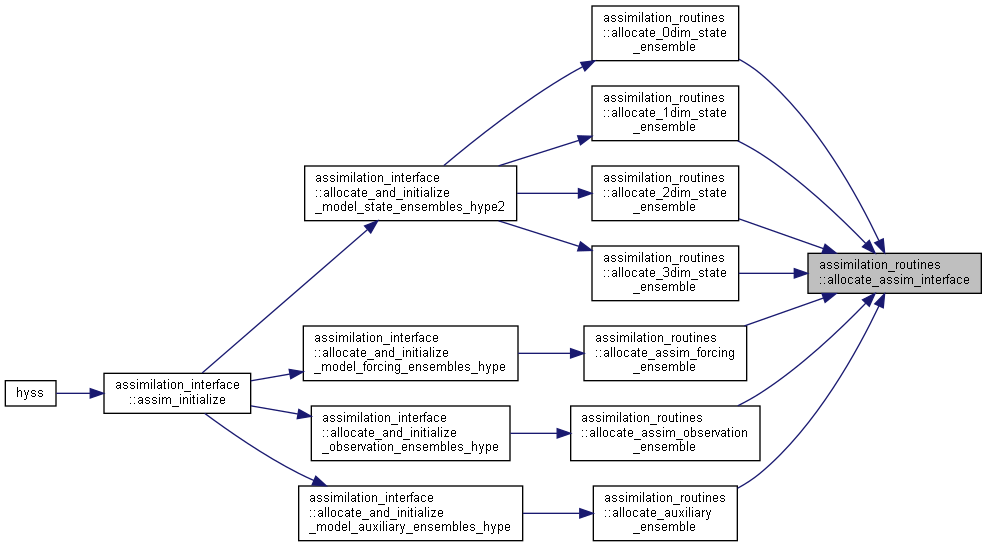
| subroutine assimilation_routines::allocate_assim_ensemble | ( | type(assim_ensemble_type) | assimVar, |
| integer | nens, | ||
| integer | nvar, | ||
| integer, intent(in) | varID, | ||
| integer, intent(in) | fileID, | ||
| character(len=*), intent(in) | bindir, | ||
| integer, intent(in) | useBinFile, | ||
| integer | locID, | ||
| integer | coordID, | ||
| real, dimension(nvar) | xini, | ||
| real | mini, | ||
| real | maxi, | ||
| logical | allocateOutput, | ||
| real | missing, | ||
| logical, intent(in) | assimilate, | ||
| logical, intent(in) | iniFromBin, | ||
| integer, intent(in) | transformation, | ||
| real, intent(in) | lambda, | ||
| real, intent(in) | epsilon, | ||
| logical, intent(in) | dxadd | ||
| ) |
Allocate and initialize an assim_ensemble_type variable.
- Parameters
-
assimvar ensemble data nens number of ensemble members nvar number of variables (model units, i.e. n:o subbasins in HYPE) [in] varid to set record for binary file [in] fileid file unit ID for direct access binary file I/O coordid ID for coordinate system for this variable xini initial values allocateoutput flag for output variable allocation missing initial output values [in] bindir path to bin-file [in] usebinfile flag for using bin-file [in] assimilate flag for including variable in assimilation [in] inifrombin flag for initializing ensemble from bin-files [in] dxadd flag for allocate and initialize perturbation field (dx)
 Here is the caller graph for this function:
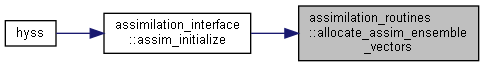
Here is the caller graph for this function:◆ allocate_assim_ensemble_vectors()
| subroutine assimilation_routines::allocate_assim_ensemble_vectors | ( | type(assim_data_type), intent(inout) | assimData, |
| integer | fid_0 | ||
| ) |
Allocate ensemble vectors needed for the data assimilation application.
- Parameters
-
[in,out] assimdata main assimilation variable containing all data fid_0 base fileunit for bin files to be used
 Here is the caller graph for this function:
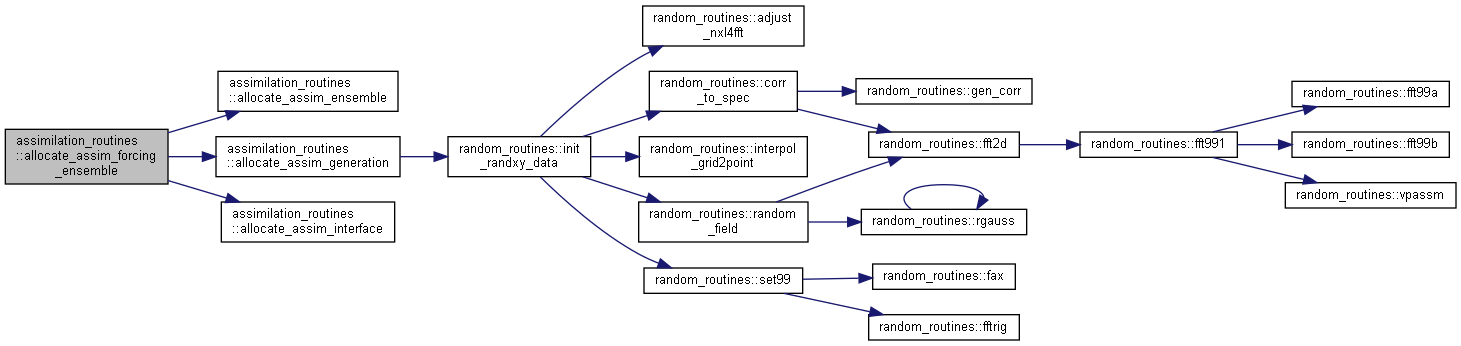
Here is the caller graph for this function:◆ allocate_assim_forcing_ensemble()
| subroutine assimilation_routines::allocate_assim_forcing_ensemble | ( | type(assim_input_ensemble_type) | assimVar, |
| integer | nens, | ||
| integer | nvar, | ||
| character(len=*) | varName, | ||
| real, dimension(nvar) | xini, | ||
| integer, intent(in) | varID, | ||
| integer | locID, | ||
| integer | coordID, | ||
| integer | fileID, | ||
| real | minimum, | ||
| real | maximum, | ||
| integer | ensgen, | ||
| real | sigma, | ||
| real | semimeta, | ||
| real | restmeta, | ||
| real | minsigma, | ||
| real | lscale, | ||
| real | gridsize, | ||
| integer | corrtype, | ||
| real, dimension(:) | xcoord, | ||
| real, dimension(:) | ycoord, | ||
| character(len=*), intent(in) | bindir, | ||
| integer, intent(in) | useBinFile, | ||
| real | missing, | ||
| logical, intent(in) | iniFromBin, | ||
| integer, intent(in) | transformation, | ||
| real, intent(in) | lambda, | ||
| real, intent(in) | epsilon, | ||
| real, intent(in) | tau, | ||
| logical, intent(in) | usedx | ||
| ) |
Allocate and initialize forcing ensemble variables.
- Parameters
-
[in] varid counter for variables in ensembles (is not all variables varID for F-variables but hardkoded) [in] bindir path to bin-file [in] usebinfile flag for using bin-file [in] inifrombin flag for initializing ensemble from bin-files
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
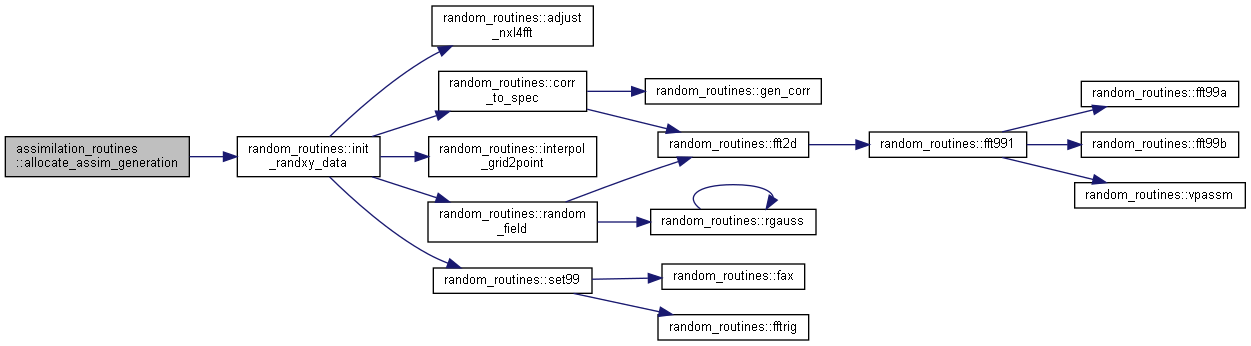
Here is the caller graph for this function:◆ allocate_assim_generation()
| subroutine assimilation_routines::allocate_assim_generation | ( | type(assim_generation_type) | assimVar, |
| integer | nvar, | ||
| integer | ensgen, | ||
| real | fixsigma, | ||
| real | semimeta, | ||
| real | restmeta, | ||
| real | minsigma, | ||
| real | lscale, | ||
| real | gridsize, | ||
| integer | corrtype, | ||
| real, dimension(:) | xcoord, | ||
| real, dimension(:) | ycoord, | ||
| real | tau | ||
| ) |
Allocate and initialize an enkf_generation_type variable.
- Parameters
-
assimvar generation_data variable nvar number of variables, ie n:o "model units" (for instance, number of sub-basins) ensgen type of ensemble generation (0 none, 1 unrestricted, 2 [min,+inf], 3 [-inf,max], 4 restricted [min,max]) fixsigma fixed standard deviation (ensgen=1) semimeta relative sigma for semi-restricted (ensgen=2,3,4, following Turner et al 2008) restmeta relative sigma for restricted (ensgen=2,3,4) minsigma minimum sigma (ensgen=2,3,4) lscale correlation length for spatially correlated perturbation gridsize 2D gridsize for spatially correlated perturbation generation corrtype spatial correlation FUNCTION option xcoord x coordinate, for spatially correlated perturbations ycoord y coordinate, for spatially correlated perturbations tau perturbation memory coefficient
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ allocate_assim_interface()
| subroutine assimilation_routines::allocate_assim_interface | ( | type(assim_interface_type) | assimVar, |
| character(len=*) | varName, | ||
| integer | varID, | ||
| integer | modID, | ||
| integer | nSubDim, | ||
| integer, dimension(:) | subDimID | ||
| ) |
Allocate and initialize a assim_interface_type variable.
- Parameters
-
assimvar interface data varname character string for model variables (used for debugging, and possibly file names and outputs) varid variable ID (id number used by interface for linking to model variables) (in HYPE it can be an outvar index, or the order in which the state variables are considered by interface) modid model ID, link to the corresponding variables used for H(X) (in HYPE: outvar index) nsubdim number of sub-dimensions (if needed, for instance lateral sub-units, vertical layers, substances, etc, in HYPE for instance SLC, substances, landuse, or slc, etc) subdimid index in the sub-dimensions
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ allocate_assim_observation_ensemble()
| subroutine assimilation_routines::allocate_assim_observation_ensemble | ( | type(assim_input_ensemble_type) | assimVar, |
| integer | nens, | ||
| integer | nvar, | ||
| character(len=*) | varName, | ||
| real, dimension(nvar) | xini, | ||
| integer | obsID, | ||
| integer | modID, | ||
| integer | coordID, | ||
| integer | fileID, | ||
| real | minimum, | ||
| real | maximum, | ||
| integer | ensgen, | ||
| real | sigma, | ||
| real | semimeta, | ||
| real | restmeta, | ||
| real | minsigma, | ||
| real | lscale, | ||
| real | gridsize, | ||
| integer | corrtype, | ||
| real, dimension(:) | xcoord, | ||
| real, dimension(:) | ycoord, | ||
| real | missing, | ||
| integer, intent(in) | transformation, | ||
| real, intent(in) | lambda, | ||
| real, intent(in) | epsilon | ||
| ) |
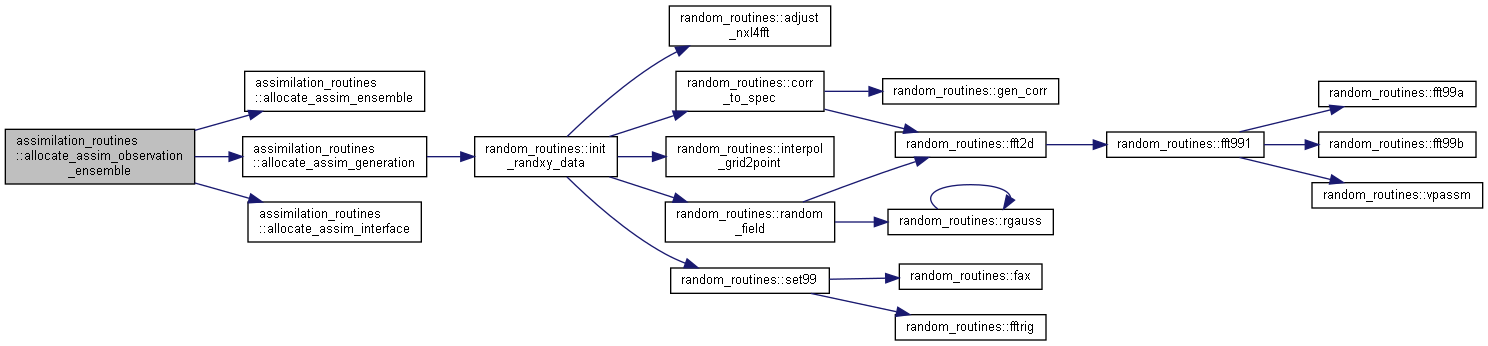
Allocate and initialize observation ensemble variables.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ allocate_auxiliary_ensemble()
| subroutine assimilation_routines::allocate_auxiliary_ensemble | ( | type(assim_state_ensemble_type) | assimVar, |
| integer | nens, | ||
| integer | nvar, | ||
| character(len=*) | varName, | ||
| real, dimension(nvar) | xini, | ||
| integer, intent(in) | varID, | ||
| integer, intent(in) | recID, | ||
| integer | locID, | ||
| integer | coordID, | ||
| integer | fileID, | ||
| character(len=*), intent(in) | bindir, | ||
| integer, intent(in) | useBinFile, | ||
| real | minimum, | ||
| real | maximum, | ||
| logical, intent(in) | assimilate, | ||
| real | missing, | ||
| integer, intent(in) | transformation, | ||
| real, intent(in) | lambda, | ||
| real, intent(in) | epsilon | ||
| ) |
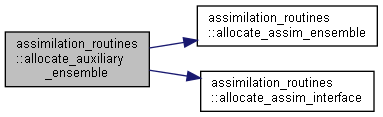
Allocate and initialize auxiliary ensemble variables; 0-dimensional.
- Parameters
-
[in] varid outvar-variable index [in] recid record of variable in bin-file [in] bindir path to bin-file [in] usebinfile flag for using bin-file [in] assimilate flag for including variable in assimilation
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ assim_backward_transform()
| subroutine assimilation_routines::assim_backward_transform | ( | integer, intent(in) | nx, |
| integer, intent(in) | ne, | ||
| real, dimension(nx,ne), intent(inout) | x, | ||
| type(assim_ensemble_type), intent(inout) | ensdata, | ||
| real, intent(in) | missing | ||
| ) |
assim_forward_transform
transformation (forward) of ensembles (observations, states, etc) to improve Gaussian nature of ensemble distributions
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ assim_checkminmax()
| subroutine assimilation_routines::assim_checkminmax | ( | integer, intent(in) | nx, |
| integer, intent(in) | ne, | ||
| real, dimension(nx,ne), intent(inout) | ensemble, | ||
| real, intent(in) | minval, | ||
| real, intent(in) | maxval, | ||
| real, intent(in) | missing | ||
| ) |
Truncates an ensemble matrix to minimum and maximum allowed values.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ assim_ensemble_statistics()
| subroutine assimilation_routines::assim_ensemble_statistics | ( | real, dimension(dim,nn) | xin, |
| integer | DIM, | ||
| integer | NN, | ||
| real, dimension(dim) | xmean, | ||
| real, dimension(dim) | xmins, | ||
| real, dimension(dim) | xmaxs, | ||
| real, dimension(dim) | xsigma, | ||
| real, dimension(dim) | xmedian, | ||
| logical | domedian, | ||
| real | missing | ||
| ) |
Calculates summary statistics for an ensemble matrix.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ assim_forward_transform()
| subroutine assimilation_routines::assim_forward_transform | ( | integer, intent(in) | nx, |
| integer, intent(in) | ne, | ||
| real, dimension(nx,ne), intent(inout) | x, | ||
| type(assim_ensemble_type), intent(inout) | ensdata, | ||
| real, intent(in) | missing | ||
| ) |
assim_forward_transform (Gaussian Anamorphosis)
transformation (forward) of ensembles (observations, states, etc) to improve Gaussian nature of ensemble distributions
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ assim_get_ensemble_data()
| subroutine assimilation_routines::assim_get_ensemble_data | ( | integer, intent(in) | NX, |
| integer, intent(in) | NE, | ||
| type(assim_ensemble_type), intent(in) | ensData, | ||
| real, dimension(nx,ne), intent(out) | x | ||
| ) |
Routine that returns the ensemble data in a matrix. If needed, the data is read from binary file. For speed, records in binary files contain all states in one ensemble member (X transformated).
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ assim_get_ensemble_data_dx()
| subroutine assimilation_routines::assim_get_ensemble_data_dx | ( | integer, intent(in) | NX, |
| integer, intent(in) | NE, | ||
| type(assim_ensemble_type), intent(in) | ensData, | ||
| real, dimension(nx,ne), intent(out) | dx | ||
| ) |
Routine that returns the perturbation field ensemble data in a matrix. If needed, the data is read from binary file. For speed, records in binary files contain all states in one ensemble member (X transformated).
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ assim_set_ensemble_data()
| subroutine assimilation_routines::assim_set_ensemble_data | ( | integer, intent(in) | NX, |
| integer, intent(in) | NE, | ||
| type(assim_ensemble_type), intent(inout) | ensData, | ||
| real, dimension(nx,ne), intent(inout) | x, | ||
| logical, intent(in) | checkMinMax, | ||
| real, intent(in) | missing | ||
| ) |
Routine that writes a matrix into an ensemble data structure. If needed, the data is written to a binary file.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ assim_set_ensemble_data_dx()
| subroutine assimilation_routines::assim_set_ensemble_data_dx | ( | integer, intent(in) | NX, |
| integer, intent(in) | NE, | ||
| type(assim_ensemble_type), intent(inout) | ensData, | ||
| real, dimension(nx,ne), intent(inout) | x, | ||
| real, dimension(nx,ne), intent(inout) | dx, | ||
| logical, intent(in) | checkMinMax, | ||
| real, intent(in) | missing | ||
| ) |
Routine that writes a matrix into an ensemble data structure. If wanted, the data is written to a binary file.
dx-version which also writes the dx field (perturbation)
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ choldc()
| subroutine assimilation_routines::choldc | ( | real, dimension(:,:), intent(inout) | a, |
| real, dimension(:), intent(out) | p, | ||
| integer | n, | ||
| logical, intent(inout) | failure | ||
| ) |
Cholesky decomposition (from Numerical Recipes, adopted by David)
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ choleskysolution()
| subroutine assimilation_routines::choleskysolution | ( | integer, intent(in) | ny, |
| integer, intent(in) | ne, | ||
| real, dimension(ny,ny), intent(inout) | P, | ||
| real, dimension(ny,ne), intent(in) | Y, | ||
| real, dimension(ny,ne), intent(out) | M, | ||
| logical, intent(inout) | failure | ||
| ) |
Cholesky solution.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ cholsl()
| subroutine assimilation_routines::cholsl | ( | real, dimension(:,:), intent(in) | a, |
| real, dimension(:), intent(in) | p, | ||
| real, dimension(:), intent(in) | b, | ||
| real, dimension(:), intent(inout) | x, | ||
| integer | n | ||
| ) |
Cholesky solver (from Numerical Recipes, adopted by David)
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ enkf_analysis_apply()
| subroutine assimilation_routines::enkf_analysis_apply | ( | integer, intent(in) | N, |
| integer, intent(in) | NX, | ||
| integer, intent(in) | ND, | ||
| real, dimension(nd,n), intent(in) | M, | ||
| real, dimension(nd,n), intent(in) | HA, | ||
| real, dimension(nx,nd), intent(in) | locCxy, | ||
| real, dimension(nx,n), intent(inout) | X | ||
| ) |
Part 2 of ensemble Kalman filter analysis.
Apply ENKF filter on a model variable ensemble using the M matrix derived in the enkf_analysis_prepare function.
EnKF analysis, following the "basic implementation" in Jan Mandel report modified to include covariance localization:
(K = Cxy .* loc_Cxy / (Cyy .* loc_Cyy + R))
The localization of Cyy is embedded in the M matrix which is prepared by the enkf_analysis_prepare routine.
The localization of Cxy is introduced in the final step of this "apply"-routine, which involves a change in the calculation of the AZ matrix compared to Mandel:
Mandel: AZ = A * (HA' * M)
Here: AZ = ((A*HA') .* loc_Cxy) * M
Matrix operations are made in an order defined by the parantheses (most inner operations first). This might have consequences on the number of operations compared to Mandel, but I have not checked that.
- Parameters
-
[in] n number of ensemble members [in] nx number of variables in ensemble (rows) [in] nd number of observations [in,out] x ensemble data [in] m inverse of (R+CXY) multiplied with innovation Y [in] ha HX deviation from mean(HX) [in] loccxy localization matrix
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
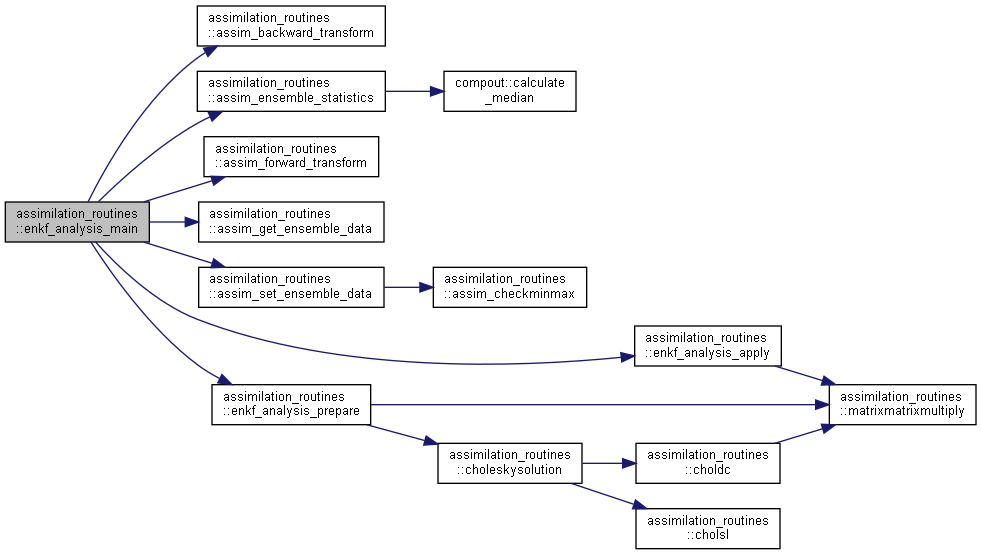
Here is the caller graph for this function:◆ enkf_analysis_main()
| subroutine assimilation_routines::enkf_analysis_main | ( | type(assim_data_type), intent(inout) | assimData | ) |
Main routine in the ensemble Kalman filter analysis.
Ensemble Kalman filter equations (Evensen) following (a) Mandel, J. Efficient implementation of the ensemble kalman filter, and (b) DeChant, C. Examining the effectiveness and robustness of sequential data assimilation methods for quantification of uncertainty in hydrological forecasting. Localization following Magnusson, Gustafsson et al (2014)
- Parameters
-
[in,out] assimdata main assimilation variable containing all data
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ enkf_analysis_prepare()
| subroutine assimilation_routines::enkf_analysis_prepare | ( | integer, intent(in) | N, |
| integer, intent(in) | ND, | ||
| real, dimension(nd,n), intent(in) | D, | ||
| real, dimension(nd,n), intent(in) | HX, | ||
| real, dimension(nd), intent(in) | R, | ||
| real, dimension(nd,nd), intent(in) | locCyy, | ||
| real, dimension(nd,n), intent(out) | M, | ||
| real, dimension(nd,n), intent(out) | Y, | ||
| real, dimension(nd,n), intent(out) | HA, | ||
| logical, intent(inout) | status | ||
| ) |
Part 1 of ensemble Kalman filter analysis.
Innovations (Y=D-HX), and inversion of variances M=1/(var(HX) + R) which is later used in part 2; the update equation:
X_analysis = X_forecast + cov(X,HX)/(var(HX)+R) * (D-HX)
- Parameters
-
[in] n number of ensemble members [in] nd number of observations [in] d observation ensemble [in] hx predicted observation ensemble [in] r observation error variance, uncorrelated [in] loccyy localization matrix for cov(HX) [out] m inverse of (R+CXY) multiplied with innovation Y [out] y innovation ensemble [out] ha HX deviation from mean(HX) [in,out] status error status, failure of cholesky
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ generate_input_ensemble()
| subroutine assimilation_routines::generate_input_ensemble | ( | integer | n, |
| type(assim_input_ensemble_type), dimension(n) | assimVar, | ||
| real | missing, | ||
| logical | usedx | ||
| ) |
General routine for Ensemble generation (forcing and observation data).
The ensemble generation is made by adding random numbers to the input data.
a) The basic assumption is that the random perturbations are gaussian with zero mean and standard deviation sigma.
b) Based on Turner et al (2008), sigma is a constant value for unrestricted variables, and a relative value for variables restricted by a minimum or a maximum or both: pertubation_unrestricted = G(0,sigma) pertubation_restricted,min = G(0,semi.meta * (X-minX)) pertubation_restricted,max = G(0,semi.meta * (maxX-X)) pertubation_restricted,minmax = G(0,rest.meta * (maxX-X)/(maxX-midX)) if X > midX
pertubation_restricted,minmax = G(0,rest.meta * (X-minX)/(midX-minX)) if X <= midX
(Turner et al(2008) also suggested that in order to get unbiased input ensembles, we need to consider two types of perturbations - systematic and random: Thus, the input x at time k for ensemble member j, x_kj = x_k0+eata_kj+chi_j, where eata_kj is regenerated every time step and chi_j is generated only once. Then, eata and chi can be generated with three different types of restrictions on variables.
However, we never implemented this part of Turner - we assume systematic error chi_j = 0.)
c) We now also take into account spatial correlation in the data, by generating spatially correlated random data using a FFT-based method (usually only on forcing)
d) Instead of the systematic error suggested by Turner et al (2008); see section in brackets under b) we introduce a memory in the perturbation (only for forcing):
pert(t) = pert(t-1) * TAU + pert * (1-TAU), where pert is generated with existing methods
for restricted and semi-restricted variables, we probably need to propagate relative perturbations instead of absolute perturbations.
 Here is the call graph for this function:
Here is the call graph for this function:◆ get_random_vector_gaussian()
| subroutine assimilation_routines::get_random_vector_gaussian | ( | integer, intent(in) | n, |
| real, intent(in) | a, | ||
| real, intent(in) | sigma, | ||
| real, dimension(:), intent(out) | r | ||
| ) |
Get an array of random gaussian values with specified mean and sigma.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ get_spatially_correlated_random_data2()
| subroutine assimilation_routines::get_spatially_correlated_random_data2 | ( | integer, intent(in) | n, |
| integer, intent(in) | nens, | ||
| type(assim_generation_type), intent(inout) | assimG, | ||
| real, dimension(n,nens), intent(out) | X, | ||
| logical | usedx | ||
| ) |
Function for spatially correlated random variable generation.
- Parameters
-
[in] n number of element (nsub usually) [in] nens number of ensembles [in,out] assimg information on generation of input data [out] x generated random matrix
Algorithm Loop over ensemble members:
- generate random numbers (with mean 0, standard deviation 1 and correlation length L) for all subbasins' location
- scale perturbations with the required sigma (standard deviation) and apply to the mean (if not perturbation is requested by USEDX
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ initialize_assim_info()
| subroutine assimilation_routines::initialize_assim_info | ( | type(assim_info_type), intent(inout) | assimInfo | ) |
Initializes the information variable for assimilation.
- Parameters
-
[in,out] assiminfo information on assimilation simulation
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ initiate_fileid_0dim_state_ensemble()
| subroutine assimilation_routines::initiate_fileid_0dim_state_ensemble | ( | type(assim_state_ensemble_type) | assimVar, |
| integer, intent(in) | nens, | ||
| integer, intent(inout) | varID, | ||
| integer, intent(in) | fileID, | ||
| integer, intent(in) | useBinFile | ||
| ) |
Initialize state ensemble variables regaring files; 0-dimensional.
- Parameters
-
[in] usebinfile flag for using bin-file
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ initiate_fileid_1dim_state_ensemble()
| subroutine assimilation_routines::initiate_fileid_1dim_state_ensemble | ( | type(assim_state_ensemble_type), dimension(:) | assimVar, |
| integer | nens, | ||
| integer | varID, | ||
| integer, dimension(:) | fileID, | ||
| integer, intent(in) | useBinFile, | ||
| integer | n1 | ||
| ) |
Initialize state ensemble variables regaring files; 1-dimensional.
- Parameters
-
[in] usebinfile flag for using bin-file
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ initiate_fileid_2dim_state_ensemble()
| subroutine assimilation_routines::initiate_fileid_2dim_state_ensemble | ( | type(assim_state_ensemble_type), dimension(:) | assimVar, |
| integer | nens, | ||
| integer | varID, | ||
| integer, dimension(:) | fileID, | ||
| integer, intent(in) | useBinfile, | ||
| integer | n1, | ||
| integer | n2 | ||
| ) |
Initialize state ensemble variables regaring files; 2-dimensional.
- Parameters
-
[in] usebinfile flag for using bin-file
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ initiate_fileid_3dim_state_ensemble()
| subroutine assimilation_routines::initiate_fileid_3dim_state_ensemble | ( | type(assim_state_ensemble_type), dimension(:) | assimVar, |
| integer | nens, | ||
| integer | varID, | ||
| integer, dimension(:) | fileID, | ||
| integer, intent(in) | useBinFile, | ||
| integer | n1, | ||
| integer | n2, | ||
| integer | n3 | ||
| ) |
Initialize state ensemble variables regaring files; 3-dimensional.
- Parameters
-
[in] usebinfile flag for using bin-files
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ initiate_fileid_assim_ensemble()
| subroutine assimilation_routines::initiate_fileid_assim_ensemble | ( | type(assim_ensemble_type) | assimVar, |
| integer, intent(in) | nens, | ||
| integer, intent(in) | varID, | ||
| integer, intent(in) | fileID, | ||
| integer, intent(in) | useBinFile, | ||
| logical, intent(in) | dxadd | ||
| ) |
Initialize file related variables of assim_ensemble_type variable.
- Parameters
-
assimvar ensemble data [in] nens number of ensemble members [in] varid to set record for binary file [in] fileid file unit ID for direct access binary file I/O [in] usebinfile flag for using bin-file [in] dxadd flag for allocate and initialize perturbation field (dx)
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ initiate_fileid_forcing_ensemble()
| subroutine assimilation_routines::initiate_fileid_forcing_ensemble | ( | type(assim_input_ensemble_type), intent(inout) | assimVar, |
| integer, intent(in) | nens, | ||
| integer, intent(in) | varID, | ||
| integer, intent(in) | fileID, | ||
| integer, intent(in) | useBinFile, | ||
| logical, intent(in) | usedx | ||
| ) |
Initialize forcing ensemble variables regaring files.
- Parameters
-
[in] varid counter for variables in forcing ensembles (is not all variables varID for F-variables but special hardkoded) [in] usebinfile flag for using bin-file
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ matrixmatrixmultiply()
| subroutine assimilation_routines::matrixmatrixmultiply | ( | real, dimension(:,:), intent(in) | mat1, |
| real, dimension(:,:), intent(in) | mat2, | ||
| real, dimension(:,:), intent(out) | matout | ||
| ) |
Subroutine for matrix multiplication.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ meanormedian_to_ensemble()
| subroutine assimilation_routines::meanormedian_to_ensemble | ( | integer, intent(in) | nx, |
| type(assim_state_ensemble_type), dimension(:) | assimX, | ||
| logical, intent(in) | meanORmedian | ||
| ) |
- Parameters
-
assimx ensemble data [in] nx number of variables [in] meanormedian flag for setting mean (T) or median (F)
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ updateensemblestatistics()
| subroutine assimilation_routines::updateensemblestatistics | ( | type(assim_data_type) | assimData, |
| real, dimension(4), intent(inout), optional | total_time | ||
| ) |
Calculates ensemble statistics for state, forcing and auxiliary ensembles.
- Parameters
-
assimdata main assimilation variable containing all data [in,out] total_time optional timing of the subroutine
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function: