Functions/Subroutines | |
| subroutine, public | load_observations (dir, odir, ns, bdate, edate, ndt, status) |
| subroutine, public | load_ascii_forcing (dir, ns, bdate, edate, ndt, status) |
| subroutine | load_one_forcing_variabel (ns, ndt, bdate, edate, forcobs, status) |
| integer function | get_forcing_index (name) |
| subroutine | load_netcdf_observations (dir, ns, bdate, edate, ndt, n_result) |
| subroutine, public | load_ascii_qx_observations (dir, ns, ndt, bdate, edate, status) |
| subroutine | load_netcdf_qx_observations (dir, ns, ndt, bdate, edate, status) |
| subroutine | save_xoreginformation (n, var2, nr) |
| subroutine, public | reset_observations (dir, mdir, odir, status) |
| subroutine, public | close_observations (odir, reset) |
| subroutine, public | get_current_forcing (idt, ns, current_date) |
| subroutine | get_one_current_forcing_data (idt, n, forcobs, current_date, current_value) |
| real function, dimension(n) | get_current_flow (idt, cd, n, no) |
| real function, dimension(no) | get_current_wst (idt, cd, no) |
| real function, dimension(no) | get_current_wst_from_xobs (readxobs, no) |
| real function, dimension(xcol) | get_current_otherobs (idt, cd) |
| real function, dimension(xorcol) | get_current_oregobs (idt, cd) |
| real function, dimension(xcgcol) | get_current_classobs (idt, cd) |
| subroutine, public | prepare_for_update (dir, locupall, locupnone, nsfile, ns, status) |
| subroutine | set_update_nutrientcorr (dim, ns, nrow, icorr, dataarray) |
| subroutine | get_current_time_pointsource (cd) |
| subroutine, public | load_basindata (dir, fdir, n, status) |
| subroutine, public | load_classdata (funit, infile, status) |
| subroutine | count_classdata (funit, infile, cddim, status) |
| subroutine, public | reload_classdata (funit, dir, filedate, status, statuscdc) |
| subroutine, public | load_geoclass (fname, geoclassdata, numslc, maxluse, maxsoil, maxrotation) |
| subroutine, public | set_model_base_configuration (basemodel, idir, idir2, dim, status) |
| subroutine, public | set_model_configuration (config, simconfig) |
| subroutine, public | initiate_model_parameters (ns_base, status) |
| subroutine, public | load_branchdata (dir, n, status) |
| subroutine | find_connected_lakebasins (ndim, basins, downbasins, upbasin) |
| subroutine, public | load_aquiferdata (dir, nsub, numaqu, status) |
| subroutine, public | load_glacierdata (dir, nsb, status) |
| subroutine | load_lakedata (funit, dir, n, lakedataid, status) |
| subroutine | load_damdata (funit, dir, n, status) |
| subroutine | load_flooddata (funit, dir, n, status) |
| subroutine | start_lakedata_table (maxcol, ncols, maxrow, nrows, str, xi, xr, iindex, rindex, ns, lakedataid) |
| subroutine, public | finish_lakedata_table (ns) |
| subroutine, public | read_and_calc_basindata (funit, infile, nskip, n, maxcol, lakedataid, status) |
| subroutine | read_diffuse_sources_file (funit, infile, nsbase, n, nskip, status) |
| subroutine | load_diffuse_sources_file (funit, dir, n, status) |
| subroutine, public | reload_diffuse_sources (funit, dir, nsbase, n, status, filedate) |
| subroutine | load_deposition (funit, dir, n, status) |
| subroutine, public | reload_deposition (funit, dir, basemodel, n, status, filedate) |
| subroutine, public | finish_atmdep_file_deposition (dir) |
| subroutine | load_rrcurvedata (funit, dir, n, status) |
| subroutine, public | load_pointsourcedata (dir, infile, n, status) |
| subroutine | read_pointsourcedata (dir, infile, nrows, status, fexist, readsubid, readpsid, readpstype, readlocation, pointsource, readdate1, readdate2) |
| subroutine, public | get_current_pointsources (dir, infile, ns, time, status) |
| logical function | turn_off_soilleakage () |
| subroutine, public | load_permanent_soilleakage (dir, n, bd, status) |
| subroutine, public | get_current_soilleakage (dir, n, time, status) |
| subroutine | find_seasonal_division_of_leakage (dir, infile, nleak, status) |
| subroutine | read_seasonal_leakage (dir, infile, n, geoid, nleak, status, leakage) |
| subroutine | read_first_monthly_leakage (dir, infile, bd, n, geoid, status, leakage) |
| subroutine | read_one_year_monthly_leakage (dir, infile, cd, n, geoid, status, leakage) |
| subroutine | load_management_data (funit, dir, n, status) |
| subroutine | load_forcing_information_data (funit, dir, ns, status) |
| subroutine | load_forcing_information_data_from_txt_file (funit, dir, ns, status) |
| subroutine | read_update_data (funit, infile, ns, nqsub, qupdsub, nqarsub, nwarsub, qarupdsub, warupdsub, qarupdar, warupdar, nwsub, wendsub, nrow, tpcorrarr, tncorrarr, tploccorrarr, tnloccorrarr, ncsub, cupdsub) |
| subroutine, public | calculate_path (ns) |
| subroutine, public | reform_inputdata_for_submodel (basemodel, ns) |
| integer function | get_subid_index (id, ns) |
| subroutine, public | load_cropdata (dir, n, status) |
| subroutine, public | get_hyss_arguments (dir, iseq, pflag) |
| subroutine, public | load_coded_info (dir, status, date1, date2, date3, skip, step, nsubst, rsmax, rsnum, readstatedate, mdir, rdir, fdir, odir, lldir, ldir, conductinibin, locupall, locupnone, subincrit) |
| subroutine | set_yesno_variable_from_line (linelen, linepos, line, errstr, onoff, status) |
| integer function | count_number_of_output (dim1, dim2, arr1, arr2, arr3, arr4, arr5, arr6, arr7) |
| subroutine | check_output_variables (dimout, n, varid, aggarea) |
| subroutine, public | set_outvar (nout, noutvar, noutvarclass) |
| subroutine | set_outvar_output (nout, noutvar, noutvarclass) |
| subroutine | set_outvar_crit (noutvar, noutvarclass) |
| subroutine, public | load_submodel_information (dir, submodelrun, basemodel, msub, status) |
| subroutine, public | set_outvar_for_variable_new (varid, areaagg, ovindex, noutvar, noutvarclass, cgname) |
| subroutine, public | prepare_subbasin_output (critbasins, status) |
| subroutine, public | load_output_regions (dir, nreg, status) |
| subroutine | check_outputfiles_for_illegal_input (status) |
| subroutine, public | load_otest (dir, name, status) |
| subroutine, public | load_parameters (dir, basemodel, status, seqflag, iens) |
| subroutine, public | load_optpar (dir) |
| subroutine | read_parameter_ensemble_task (dir, taskas, taskbs) |
| subroutine | calculate_special_optpar_parameters (dir, velindex, dampindex, rivvel, damp) |
| subroutine | calculate_special_partxt_parameters (dir, nruns, velindex, dampindex, rivvel, damp) |
| subroutine, public | initiate_output_routines () |
| subroutine, public | set_outvar_test_information (n) |
| subroutine, public | count_ensemble_simulations (use_bestsims, use_allsim, nens) |
| subroutine | count_parfile_simulations (nens) |
| subroutine, public | load_ensemble_simulations (dir, fname, numpar, nperf, numcrit, nsim, parameters) |
| subroutine | read_simulation_perf_and_par (funit, i, mpar, numpar, nperf, numcrit, optcrit, performance, parameters, status) |
Detailed Description
Read and process model input from files, both static data and forcing.
Procedures for observations, and for reading and preparing input data.
Function/Subroutine Documentation
◆ calculate_path()
| subroutine, public datamodule::calculate_path | ( | integer, intent(in) | ns | ) |
Calculate the coupling between subbasins by index from the existing coupling by subid. In addition set the flag for upstream lakebasins flowing to downstream lakebasin and branches that are secondary outlets of lakebasin lake.
Consequences Module modvar variables path, branchdata and branchindex may be allocated and set. Module modvar variables pathsubid and branchsubid is deallocated.
- Parameters
-
[in] ns Number of subbasins (of submodel)
Algorithm
Set main path and groundwater flow path
Check for connected lakebasins in main flow path
Set branch path and data, if present
Check for connected lakebasins in branched flow path
Check for secondary outlet of lakebasins in branched flow path
Find subbasin index that outlet originated from
◆ calculate_special_optpar_parameters()
|
private |
Reads parameters to be calibrated to find maximum dimension of river states.
- Parameters
-
[in] dir file directory [in] velindex index of rivvel in modparid [in] dampindex index of damp in modparid [in,out] rivvel lower river velocity boundary [in,out] damp lower damp boundary
◆ calculate_special_partxt_parameters()
|
private |
Reads parameters of ensemble simulation to find maximum dimension of river states.
- Parameters
-
[in] dir file directory [in] nruns number of ensembles [in] velindex index of rivvel in modparid [in] dampindex index of damp in modparid [in,out] rivvel lower river velocity boundary [in,out] damp lower damp boundary
Check whole ensemble's par files
Read each parameter from file
Find rivvel and damp values
◆ check_output_variables()
|
private |
Checks for incompatible output.
- Parameters
-
[in] dimout dimension of variable arrays [in,out] n number of output variables of this type [in,out] varid variables for output [in,out] aggarea area aggregation type of variable (subbasin=0,upstream=1,region=2)
Algorithm
Check outvar information for non-calculatable upstream variables
Change output variable information
Set output
◆ check_outputfiles_for_illegal_input()
|
private |
Check output files for illegal variables.
- Parameters
-
[out] status status returned from function
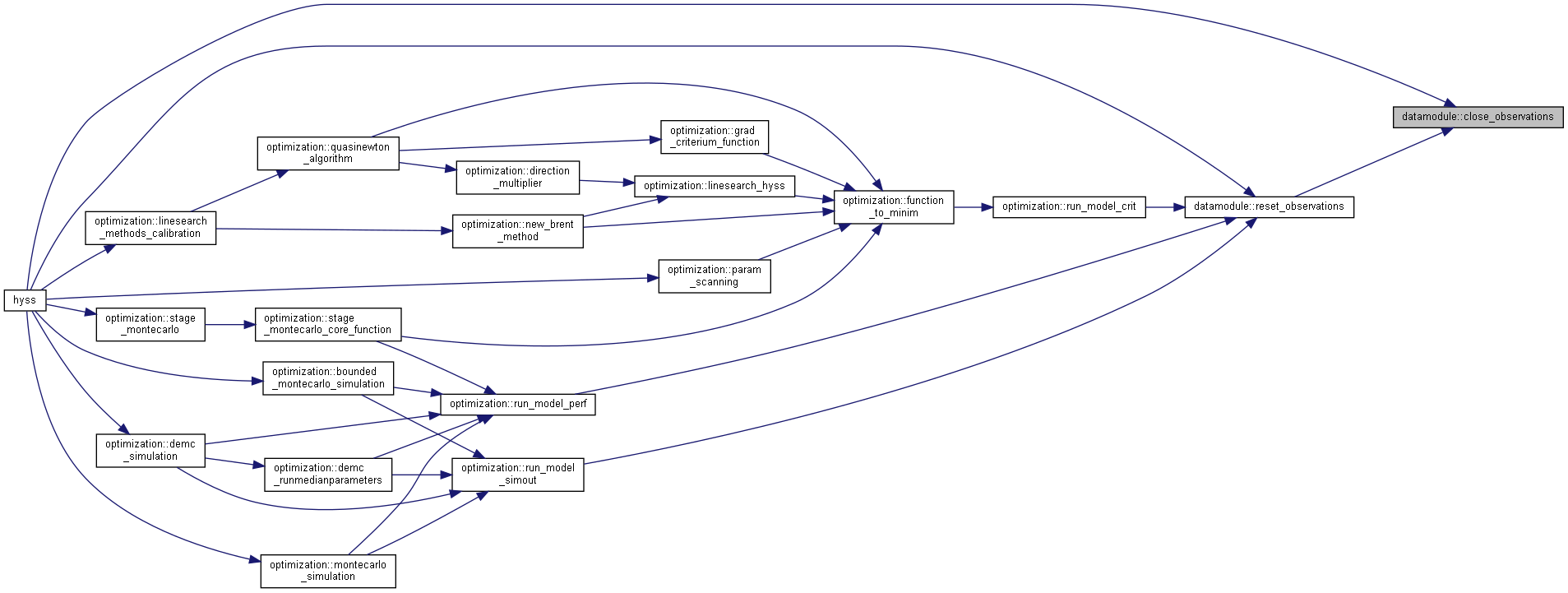
◆ close_observations()
| subroutine, public datamodule::close_observations | ( | character(len=*), intent(in) | odir, |
| logical, intent(in), optional | reset ) |
Close files with observations used for forcing and calibration.
- Parameters
-
[in] odir File directory (otherobsdir for Xobs) [in] reset flag that subroutine is used for reset
◆ count_classdata()
|
private |
Gets the information about number of classes, land uses, soil types and crop rotation groups from a ClassData.txt.
- Parameters
-
[in] funit Unit for file [in] infile Name of characteristics file to be read [out] cddim Highest index of classes, landuse, soil type and crop rotation group in file [out] status File found and read
Algoritm
Count number of classes (actually all rows)
Allocate variable for holding class information
Read information of ClassData.txt
- Skip comment lines in the beginning of the file
- Read column headings
- Read class information (requires slc in order in file NOT)
◆ count_ensemble_simulations()
| subroutine, public datamodule::count_ensemble_simulations | ( | logical, intent(in) | use_bestsims, |
| logical, intent(in) | use_allsim, | ||
| integer, intent(out) | nens ) |
Count number of parameter sets used for ensemble simulation.
- Parameters
-
[in] use_bestsims Task count bestsims.txt [in] use_allsim Task count allsim.txt [out] nens Number of parameter ensembles
Algorithm
Find file with parameter set(s)
If file with parameter ensemble sets
- Open file and skip heading
- Read number of lines
- End routine
◆ count_number_of_output()
|
private |
Count number of outputs / output types and output periods.
Consequences Module worldvar variable noutput is set by return value
- Parameters
-
[in] dim1 dimension of array (meanperiod) [in] dim2 dimension of array (groups) [in] arr1 array with number of variables for output of type 1 [in] arr2 array with number of variables for output of type 2 [in] arr3 array with number of variables for output of type 3 [in] arr4 array with number of variables for output of type 4 [in] arr5 array with number of variables for output of type 5 or 6 [in] arr6 array with number of variables for output of type 5 or 6 [in] arr7 array with number of variables for output of type 7
Algorithm
Count number of outputs with set variables, i.e. used for this simulation
◆ count_parfile_simulations()
|
private |
Count number of parameter files ensemble simulation.
- Parameters
-
[out] nens Number of parameter ensembles
Algorithm
Find file with parameter set(s)
◆ find_connected_lakebasins()
|
private |
Check for connected lakebasins.
- Parameters
-
[in] ndim Number of subbasins to be checked [in] basins Subid of all subbasins to be checked; basin(i)subid, branchsubid(i)source [in] downbasins Subid of their downstream subbasins; pathsubid(i)main or branchsubid(i)branch [out] upbasin Flag for lakebasins upstream of other lakebasins
Check for connected lakebasins
◆ find_seasonal_division_of_leakage()
|
private |
Checks a file for seasonal leakage for the seasonal division of the file The file can have seasonality as (daily or) monthly values or be constant.
- Parameters
-
[in] dir File directory [in] infile Name of leakage file [out] nleak Seasonal division; number of values to be read for each subbasin (1, 12 or 366) [out] status Error status
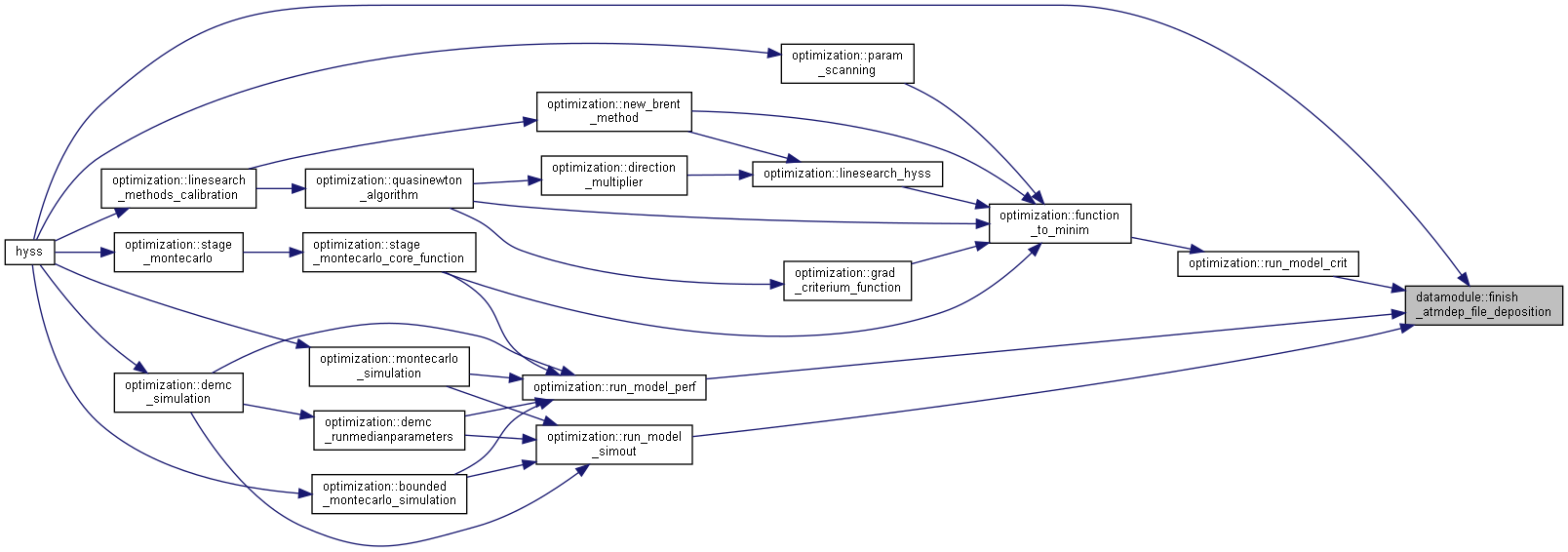
◆ finish_atmdep_file_deposition()
| subroutine, public datamodule::finish_atmdep_file_deposition | ( | character (len=*), intent(in) | dir | ) |
Set deposition to model specified values if no AtmdepData.txt file is used.
- Parameters
-
[in] dir File directory
Algorithm
Check for file
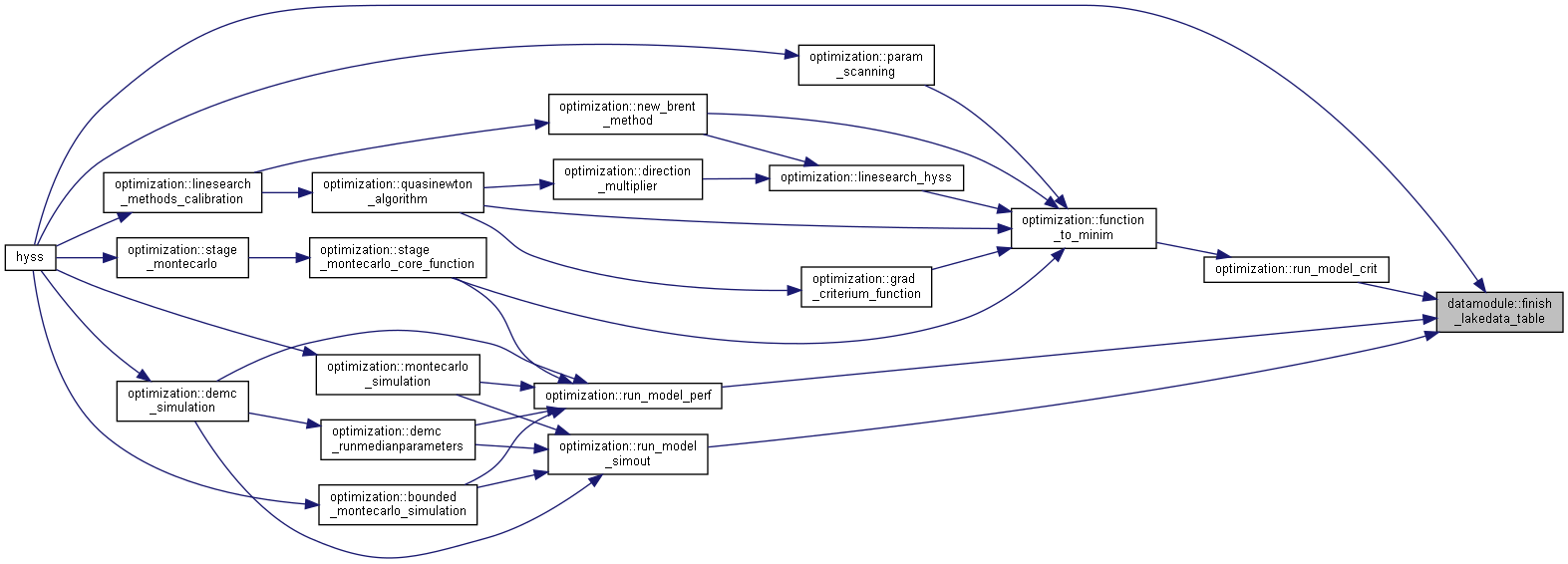
◆ finish_lakedata_table()
| subroutine, public datamodule::finish_lakedata_table | ( | integer, intent(in) | ns | ) |
Set lakedatapar values to those specified in HYPE parameter variables.
- Parameters
-
[in] ns number of subbasins
Algorithm
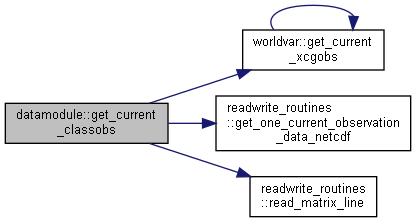
◆ get_current_classobs()
|
private |
Get current value of class observations from files or memory.
- Parameters
-
[in] idt current time step [in] cd current date
- Returns
- current observations
◆ get_current_flow()
|
private |
Get current discharge from file or memory.
- Parameters
-
[in] idt current time step [in] cd current date [in] n number of subbasins [in] no number of observation stations
- Returns
- current flow
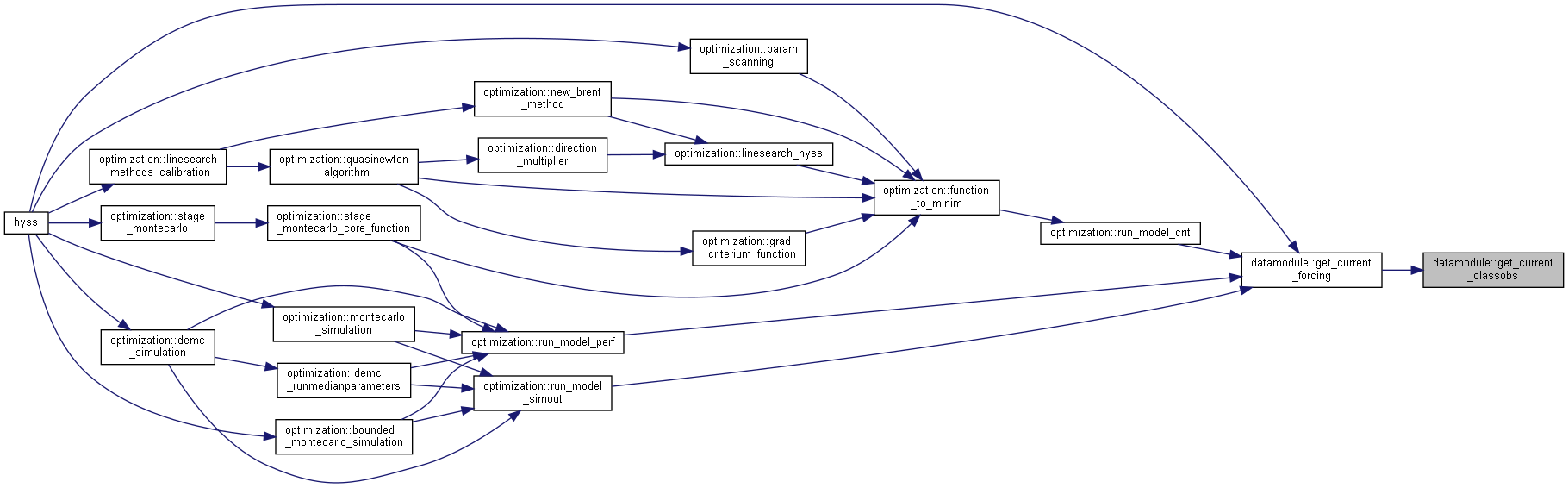
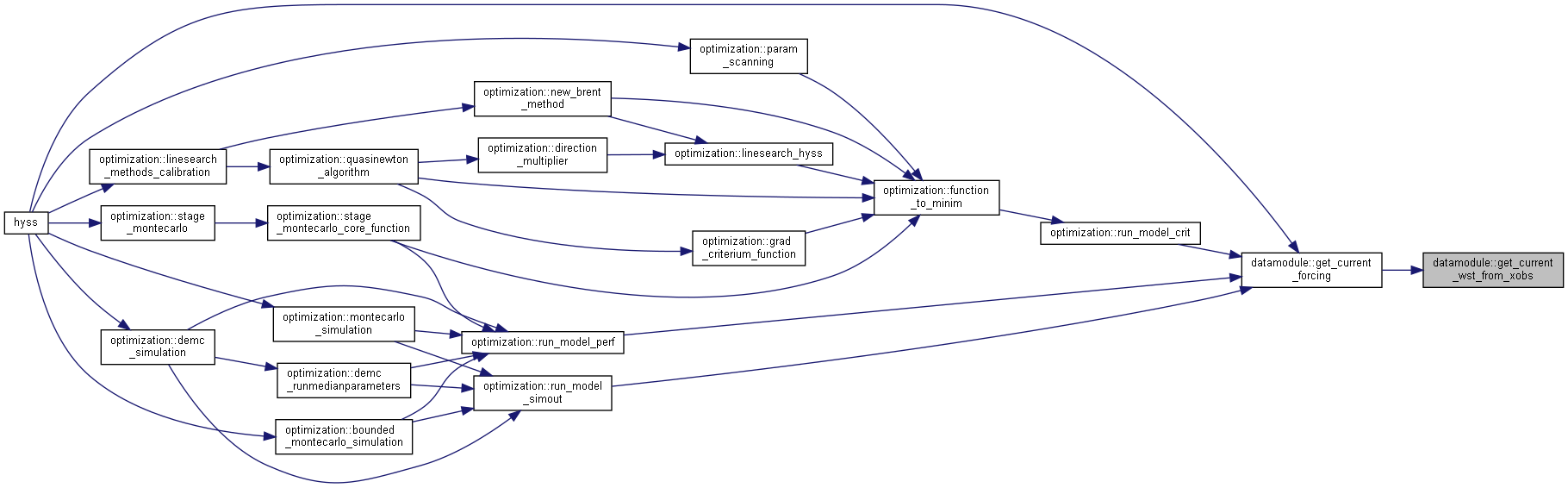
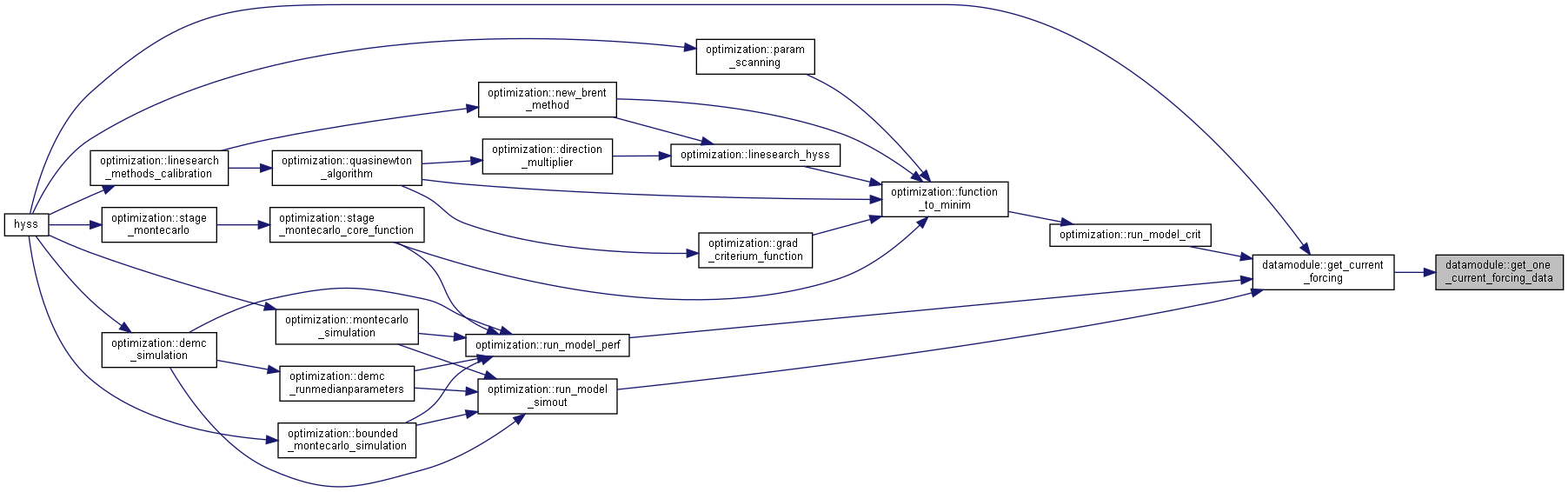
◆ get_current_forcing()
| subroutine, public datamodule::get_current_forcing | ( | integer, intent(in) | idt, |
| integer, intent(in) | ns, | ||
| type(datetype), intent(out) | current_date ) |
Get current forcing from file or memory.
- Parameters
-
[in] idt current timestep index [in] ns number of subbasins [out] current_date current date
◆ get_current_oregobs()
|
private |
Get current value of other observations from file or memory.
- Parameters
-
[in] idt current time step [in] cd current date
- Returns
- current observations
◆ get_current_otherobs()
|
private |
Get current value of other observations from file or memory.
- Parameters
-
[in] idt current time step [in] cd current date
- Returns
- current observations
◆ get_current_pointsources()
| subroutine, public datamodule::get_current_pointsources | ( | character (len=*), intent(in) | dir, |
| character (len=*), intent(in) | infile, | ||
| integer, intent(in) | ns, | ||
| type(datetype), intent(in) | time, | ||
| integer, intent(out) | status ) |
Gets the information about temporary point sources from PointSourceData.txt. Recalculate into form suitable for model.
Consequences Module modvar variable load and t1load are changed
Reference ModelDescription Chapter 5 (Point sources)
- Parameters
-
[in] dir File directory [in] infile Name of pointsource file to be read (PointSourceData.txt) [in] ns Number of subbasins (submodel) [in] time Current time (e.g. 2005-01-26 18:00) [out] status Error status
◆ get_current_soilleakage()
| subroutine, public datamodule::get_current_soilleakage | ( | character (len=*), intent(in) | dir, |
| integer, intent(in) | n, | ||
| type(datetype), intent(in) | time, | ||
| integer, intent(out) | status ) |
Gets the information about nutrient soil leakage from file.
Alternative 3: Soil nutrient leakage load is read from file. A simlified soil routine is used. Alternative 6: Soil nutrient leakage load and conc is read from files.
Consequences Module modvar variable soilleak is set.
Reference ModelDescription Chapter Nitrogen and phosphorus in land routines (Nutrient soil leakage from outer source)
- Parameters
-
[in] dir File directory [in] n Number of subbasins, submodel [in] time Current time (e.g. 2005-01-26 18:00) [out] status Error status
Algorithm
Load current soilleakage load from files
- Allocate and initiate variables
- Read data files
Load current soilleakage from files
- Allocate and initiate variables
- Read data files
◆ get_current_time_pointsource()
|
private |
Get current value of time series pointsource from file and set modvar variable psload.
- Parameters
-
[in] cd current date
◆ get_current_wst()
|
private |
Get current water level from file or memory.
- Parameters
-
[in] idt current time step [in] cd current date [in] no number of observation stations
- Returns
- current water stage of lake
◆ get_current_wst_from_xobs()
|
private |
Get current water level from xobsi array.
- Parameters
-
[in] readxobs current xobs observations [in] no number of observation stations
- Returns
- current water stage of lake
◆ get_forcing_index()
|
private |
Find index of forcing based on filename.
◆ get_hyss_arguments()
| subroutine, public datamodule::get_hyss_arguments | ( | character(len=maxcharpath), intent(out) | dir, |
| integer, intent(out) | iseq, | ||
| logical, intent(out) | pflag ) |
Get command line argument: directory of info.txt and simulation sequence, or read them from filedir.txt (old variant).
- Parameters
-
[out] dir Directory for information about simulation (infodir) [out] iseq Sequence number (simsequence) [out] pflag Sequence number to be used on par-file (parseq)
Algorithm
If no command line argument, read information from filedir.txt
Get command line argument
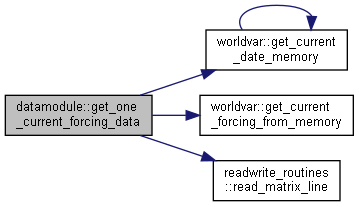
◆ get_one_current_forcing_data()
|
private |
Get current forcing data for one variable for selected subbasins from file or memory.
- Parameters
-
[in] idt current time step [in] n number of subbasins [in] forcobs data/information for one forcing variable [out] current_date current date [out] current_value current value of the forcing variable
◆ get_subid_index()
|
private |
Finds the corresponding index for a subid.
- Parameters
-
[in] id subid [in] ns number of subbasins
Algorithm
Loop through subbasin id: to find the index of the current one.
◆ initiate_model_parameters()
| subroutine, public datamodule::initiate_model_parameters | ( | integer, intent(in) | ns_base, |
| integer, intent(out) | status ) |
Allocate model parameters and set their region division coupling.
Consequences Module modvar arrays related to parameters are allocated and set.
- Parameters
-
[in] ns_base number of subbasin [out] status error status of subroutine
◆ initiate_output_routines()
| subroutine, public datamodule::initiate_output_routines |
Initiate accumulation variables for printout.
Consequences Module worldvar variables maptime,
tmap, output, and accload are set.
◆ load_aquiferdata()
| subroutine, public datamodule::load_aquiferdata | ( | character (len=*), intent(in) | dir, |
| integer, intent(in) | nsub, | ||
| integer, intent(out) | numaqu, | ||
| integer, intent(out) | status ) |
Get the information about aquifers from AquiferData.txt.
Consequences Module modvar variable aquifer and pathsubid will be set. Module modvar varible nregions may be increased.
- Parameters
-
[in] dir File directory [in] nsub Number of subbasins (basemodel) [out] numaqu Number of aquifers [out] status Error status
◆ load_ascii_forcing()
| subroutine, public datamodule::load_ascii_forcing | ( | character(len=*), intent(in) | dir, |
| integer, intent(in) | ns, | ||
| type(datetype), intent(in) | bdate, | ||
| type(datetype), intent(in) | edate, | ||
| integer, intent(in) | ndt, | ||
| integer, intent(out) | status ) |
Reads forcing data from file. Also checks data.
Consequences Module worldvar variables forcingdata and dates may change. Module modvar variables preci, tempi, snowfraci, shortwavei, windi, humidi, tmini, tmaxi, uwindi, vwindi may be allocated.
- Parameters
-
[in] dir File directory (forcingdir) [in] ns Number of subbasins, basemodel [in] bdate Begin simulation date=bdate [in] edate End simulation date=sdate [in] ndt Number of timesteps in simulation [out] status Error number
◆ load_ascii_qx_observations()
| subroutine, public datamodule::load_ascii_qx_observations | ( | character(len=*), intent(in) | dir, |
| integer, intent(in) | ns, | ||
| integer, intent(in) | ndt, | ||
| type(datetype), intent(in) | bdate, | ||
| type(datetype), intent(in) | edate, | ||
| integer, intent(out) | status ) |
Reads Qobs, Wobs, Xobs and Xoregobs data values in file.
Consequences Module worldvar variables qobs, wobs, xobs, xoregobs, xcol, xorcol, qobsdata, wobsindex, wobsindex, numwobsstn, bwdate, ewdate, bxdate, exdate, bxrdate and exrdate may change. Module modvar variables qobsi, xobsi, wobsi and xoregobsi may be allocated, and xobsindex and xoregindex may change.
- Parameters
-
[in] dir File directory (otherobsdir, earlier forcingdir) [in] ns Number of subbasins, basemodel [in] ndt Number of timesteps in simulation [in] bdate Begin simulation date [in] edate End simulation date [out] status Error number
◆ load_basindata()
| subroutine, public datamodule::load_basindata | ( | character(len=*), intent(in) | dir, |
| character(len=*), intent(in) | fdir, | ||
| integer, intent(out) | n, | ||
| integer, intent(out) | status ) |
Gets the basic information about soil-landuse-classes and subbasins from GeoClass and GeoData. Additional geographical data for lakes, dams and floodplains are also collected as well as data on water management (e.g. irrigation) and forcing data location.
Consequences Module worldvar variables atmdepvegreset may be set. Module modvar variables basin, classdata, wetland, nclass, nluse, nsoil, modeloption, conductdamconstruction, conductriverwetland and numrotations are set.
- Parameters
-
[in] dir File directory (model files) [in] fdir File directory (forcing data) [out] n Number of subbasins [out] status Error status
Algorithm
Read the class characteristics; either from ClassData or GeoClass
Count data in GeoData.txt
Initiate subbasin data and deposition and diffuse source
Read the description of the basins (GeoData.txt)
Read irrigation information of the basins (MgmtData.txt)
Read observation coupling and elevation from separate file (ForcKey.txt)
Read additional description of important lakes (LakeData.txt) (possibly overwrite GeoData information)
Read additional description of important dams (DamData.txt)
Read additional description of flooding areas (FloodData.txt)
Read additional description of river water level (RRCurveData.txt, RiverRatingCurveData.txt)
Read non point (diffuse) sources
Read atmospheric deposition sources
◆ load_branchdata()
| subroutine, public datamodule::load_branchdata | ( | character (len=*), intent(in) | dir, |
| integer, intent(in) | n, | ||
| integer, intent(out) | status ) |
Get the information about branches from BranchData.txt.
Consequences Module modvar variable branchsubid will be set
- Parameters
-
[in] dir File directory [in] n Number of subbasins (basemodel) [out] status Error status
Algorithm
Check if file exist
Allocate data holding arrays
Read BranchData-file
Initiate branch data
Save loaded branch data temporary
Check for illegal combinations of branch data and other illegal values
◆ load_classdata()
| subroutine, public datamodule::load_classdata | ( | integer, intent(in) | funit, |
| character (len=*), intent(in) | infile, | ||
| integer, intent(out) | status ) |
Gets the information about classes from ClassData.txt Reads the file with column headings.
Consequences Module variables will be allocated and set
- Parameters
-
[in] funit Unit for file [in] infile Name of characteristics file to be read [out] status File found and read
Algoritm
Check for file.
Count number of classes, land uses, soil types and rotationgroups. Set dimension variables.
Allocate variables for holding class information
Read information of ClassData.txt
- Skip comment lines in the beginning of the file
- Read column headings, and get number of columns
- Read class information (requires slc in order in file NOT)
Sort class information
◆ load_coded_info()
| subroutine, public datamodule::load_coded_info | ( | character(len=maxcharpath), intent(in) | dir, |
| integer, intent(out) | status, | ||
| type(datetype), intent(out) | date1, | ||
| type(datetype), intent(out) | date2, | ||
| type(datetype), intent(out) | date3, | ||
| integer, intent(out) | skip, | ||
| integer, intent(out) | step, | ||
| integer, intent(out) | nsubst, | ||
| integer, intent(in) | rsmax, | ||
| integer, intent(out) | rsnum, | ||
| type(datetype), dimension(rsmax), intent(out) | readstatedate, | ||
| character(len=maxcharpath), intent(out) | mdir, | ||
| character(len=maxcharpath), intent(out) | rdir, | ||
| character(len=maxcharpath), intent(out) | fdir, | ||
| character(len=maxcharpath), intent(out) | odir, | ||
| character(len=maxcharpath), intent(out) | lldir, | ||
| character(len=maxcharpath), intent(out) | ldir, | ||
| logical, intent(out) | conductinibin, | ||
| logical, dimension(dim_update), intent(out) | locupall, | ||
| logical, dimension(dim_update), intent(out) | locupnone, | ||
| integer, dimension(:), intent(inout), allocatable | subincrit ) |
Consequences The following worldvar module variables is set: readdaily, readobsid, readpstime, writeload, accload, writematlab, weightsub,
usestop84, ncrit, readformat, readoutregion, simsubmodel, resultseq, steplen, forcingdata, indtacheckonoff, indatachecklevel, doassimilation, resetstate, outstate, instate. The following modvar module variables is set: index for substances, simulate, simulatesubstances, conduct, conductxoms, conductwb, conductload, conductregest, conductwarnon, classgroup2, soiliniwet and modeloption.
- Parameters
-
[in] dir File directory [out] status Status of subroutine [out] date1 Begin date of simulation [out] date2 End date of simulation [out] date3 Begin date for criteria/output date [out] skip Length of warm-up period [out] step Length of simulation period [out] nsubst Number of substances simulated [in] rsmax Dimension dates for reading soil state [out] rsnum Number of dates for reading soil state [out] readstatedate Dates for reading soil state variables [out] mdir File directory model [out] rdir File directory result [out] fdir File directory forcing [out] odir File directory other observation (evaluation) [out] lldir File directory load leak [out] ldir File directory log [out] conductinibin Flag for reading of previously saved ensemble of starting state [out] locupall Flag for update for all stations [out] locupnone Flag for update on no stations [in,out] subincrit Subbasins to be included in criteria calculations
◆ load_cropdata()
| subroutine, public datamodule::load_cropdata | ( | character (len=*), intent(in) | dir, |
| integer, intent(out) | n, | ||
| integer, intent(out) | status ) |
Reads information about crops from file.
Consequences Module modvar variables cropdata, cropirrdata and cropdataindex may be allocated and set.
- Parameters
-
[in] dir File directory [out] n Number of rows in file [out] status Error status
Algorithm
Check if file exist and if it is required
Count number of crops and data columns
Allocate and initiate cropdata variables
Read column headings from file
Code variables for easy finding of variable type
Read the data of the crops to matrix, first column is string which is skipped
Set cropdata variables according to matrix with read data
Check if cropdata is needed?
Calculate index variable to find crops
◆ load_damdata()
|
private |
Collects the information about dams from DamData.txt.
Consequences Module modvar variables dam, damindex may be allocated and set. Module modvar variable basin and conductdamconstruction may be changed (lake_depth).
- Parameters
-
[in] funit File unit, temporary file open [in] dir File directory [in] n Number of subbasins (base model) [out] status Error status
◆ load_deposition()
|
private |
Gets the information about atmospheric deposition from AtmdepData.
Consequences Module variables deposition,deposition2 may be allocated and set
- Parameters
-
[in] funit Unit for file [in] dir File directory [in] n Number of subbasins (in GeoData) [out] status error status
Check for substance simulation
Check for file and return if no file
◆ load_diffuse_sources_file()
|
private |
Gets the information about diffuse sources for subbasins from NonPointSourceData.txt.
- Parameters
-
[in] funit Unit for file [in] dir File directory [in] n Number of subbasins (in GeoData) [out] status Error status
◆ load_ensemble_simulations()
| subroutine, public datamodule::load_ensemble_simulations | ( | character(len=*), intent(in) | dir, |
| character(len=*), intent(in) | fname, | ||
| integer, intent(in) | numpar, | ||
| integer, intent(in) | nperf, | ||
| integer, intent(in) | numcrit, | ||
| integer, intent(in) | nsim, | ||
| real, dimension(nsim,numpar), intent(inout) | parameters ) |
Load parameters used for ensemble simulation from file.
- Parameters
-
[in] dir File directory [in] fname File name [in] numpar Number of optimised parameters [in] nperf Number of performance measures [in] numcrit Number of optimised variables [in] nsim Number of simulations to be saved [in,out] parameters Parameter values for simulations
◆ load_flooddata()
|
private |
Collects the information about flooding areas from FloodData.txt.
Consequences Module modvar variables conduct,flooding,floodindex may be allocated and set.
- Parameters
-
[in] funit File unit, temporary file open [in] dir File directory [in] n Number of subbasins (base model) [out] status Error status
◆ load_forcing_information_data()
|
private |
Gets information about forcing data coupling to subbasin and other information on forcing.
Consequences Module worldvar variable forcingdata's stationid are set. Module modvar variable tobselevation is allocated and set (in subroutine).
- Parameters
-
[in] funit Unit number for file [in] dir File directory [in] ns Number of subbasins (base model) [out] status Error status
◆ load_forcing_information_data_from_txt_file()
|
private |
Gets information about forcing data coupling to subbasin and other information from ForcKey.txt.
Consequences Module worldvar variable focringdata may change. Module modvar variable tobselevation is allocated and set.
- Parameters
-
[in] funit Unit number for file [in] dir File directory [in] ns Number of subbasins (base model) [out] status Error status
◆ load_geoclass()
| subroutine, public datamodule::load_geoclass | ( | character(len=*), intent(in) | fname, |
| type(classtype), dimension(:), intent(inout), allocatable | geoclassdata, | ||
| integer, intent(out) | numslc, | ||
| integer, intent(out) | maxluse, | ||
| integer, intent(out) | maxsoil, | ||
| integer, intent(out) | maxrotation ) |
Read class characteristics file GeoClass.txt.
Consequences Module modvar variables classdata, classgroup, nclass, nluse, nsoil, numrotations and vegtypereset will be allocated and changed by the argument output of this subroutine. Module modvar variables for special classes will be set by a subroutine call.
- Parameters
-
[in] fname File path/name [in,out] geoclassdata class characteristics [out] numslc Number of land-soil use combination in GeoClass.txt [out] maxluse Number of landuses [out] maxsoil Number of soiltypes [out] maxrotation Number of groups of classes that exchange crops between years
Algorithm
Read the file with key to class characteristics (GeoClass.txt)
Set read data to class characteristic structure
Find coded classes
◆ load_glacierdata()
| subroutine, public datamodule::load_glacierdata | ( | character (len=*), intent(in) | dir, |
| integer, intent(in) | nsb, | ||
| integer, intent(out) | status ) |
Get the information about glaciers from GlacierData.txt.
Consequences Module modvar variables glacier and glacierindex will be set.
- Parameters
-
[in] dir File directory [in] nsb Number of subbasins (basemodel) [out] status Error status
◆ load_lakedata()
|
private |
Collects the information about special lakes from LakeData.txt.
Consequences Module modvar variables elake, slake, lakebasin, lakeindex, lakebasinindex may be allocated and set. Module modvar variables basin and conductdamconstruction may be changed.
- Parameters
-
[in] funit File unit, temporary file open [in] dir File directory [in] n Number of subbasins (base model) [in] lakedataid lakedataid from GeoData [out] status Error status
◆ load_management_data()
|
private |
Gets the information about irrigation and water transfer from MgmtData.
- Parameters
-
[in] funit Unit for file [in] dir File directory [in] n Number of subbasins [out] status Error status
◆ load_netcdf_observations()
|
private |
- Parameters
-
[in] dir File directory [in] ns Number of subbasins, basemodel [in] bdate Begin simulation date [in] edate End simulation date [in] ndt Number of timesteps [out] n_result Error number
◆ load_netcdf_qx_observations()
|
private |
Reads Qobs.nc, NNNNobs.nc, RGNNNNobs.nc data values in file.
Consequences Module worldvar variables qobs, xobs, xoregobs, xcol, xorcol, qobsdata, wobsindex, numwobsstn, bxdate, exdate, bxrdate and exrdate may change. Module modvar variables qobsi, xobsi, wobsi and xoregobsi may be allocated, and xobsindex and xoregindex may change.
- Parameters
-
[in] dir File directory (forcingdir) [in] ns Number of subbasins, basemodel [in] ndt Number of timesteps in simulation [in] bdate Begin simulation date [in] edate End simulation date [out] status Error number
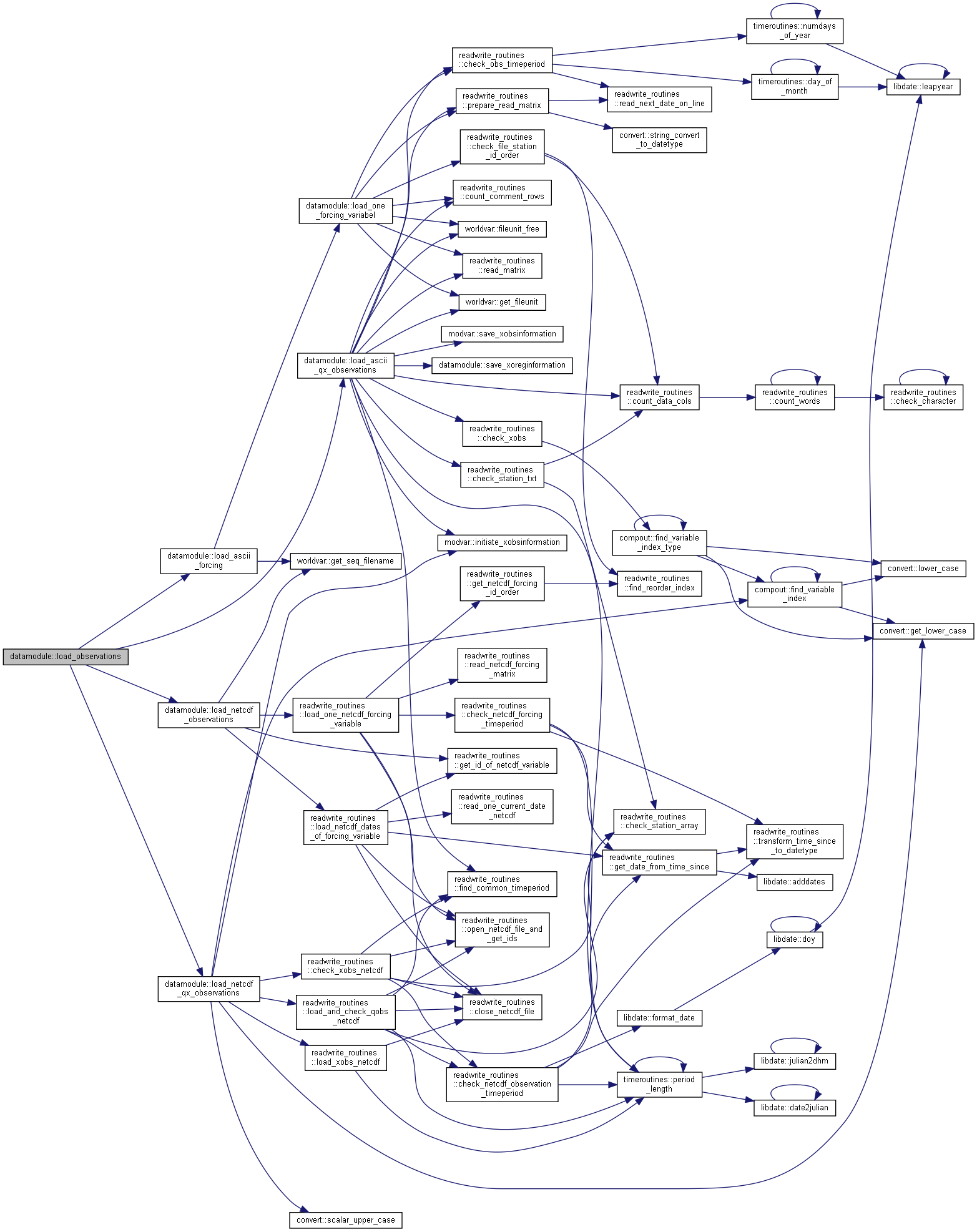
◆ load_observations()
| subroutine, public datamodule::load_observations | ( | character(len=*), intent(in) | dir, |
| character(len=*), intent(in) | odir, | ||
| integer, intent(in) | ns, | ||
| type(datetype), intent(in) | bdate, | ||
| type(datetype), intent(in) | edate, | ||
| integer, intent(in) | ndt, | ||
| integer, intent(out) | status ) |
Load forcing data files and other observations into the program.
- Parameters
-
[in] dir File directory (forcingdir) [in] odir File directory (otherobsdir) [in] ns Number of subbasins, basemodel [in] bdate Begin time for simulation [in] edate End time for simulation [in] ndt Number of timesteps [out] status Error status of subroutine
Algorithm
Load observations from files
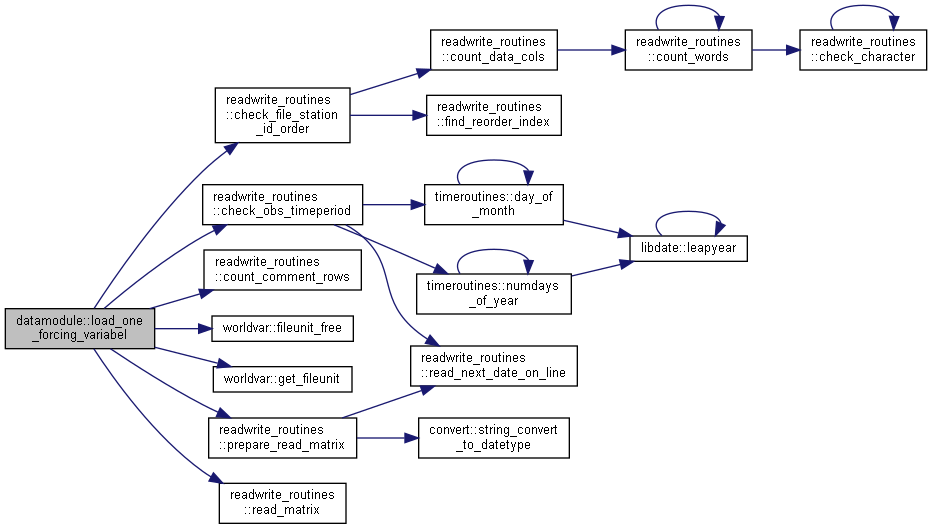
◆ load_one_forcing_variabel()
|
private |
Load one observation forcing file; check file, saves data or prepare to read continously.
- Parameters
-
[in] ns Number of subbasins, basemodel [in] ndt Number of timesteps in simulation [in] bdate Begin simulation date=bdate [in] edate End simulation date=sdate [in,out] forcobs Forcing data information and values [out] status Error number
◆ load_optpar()
| subroutine, public datamodule::load_optpar | ( | character(len=*), intent(in) | dir | ) |
Reads file with intructions on calibration and parameters to be calibrated and saves them in worldvar-arrays. Also reads file for instructions and parameters for parameter ensemble simulation.
Consequences Module worldvar variables optparmin, optparmax, optparprecision, optparid, parindex, optim, dimpar, numoptimpar will be allocated and set.
- Parameters
-
[in] dir File directory
Algorithm
Check for file
Initiation of variables for optimization parameters
Open and read simulation settings from optpar.txt
Set tasks for calibration
Set tasks for parameter ensemble simulation
Continue to read parameters
Sort min and max values
Set and check worldvar numoptimpar = amount of model parameters to be optimized
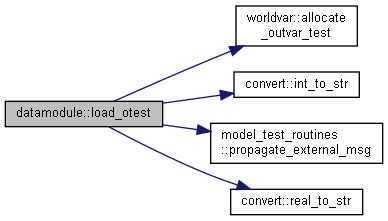
◆ load_otest()
| subroutine, public datamodule::load_otest | ( | character(len=*), intent(in) | dir, |
| character(len=*), intent(in) | name, | ||
| integer, intent(out) | status ) |
Read output test values from file.
- Parameters
-
[in] dir File directory [in] name File name [out] status Error status of subroutine
Open file and read heading
Read parameter values from file
◆ load_output_regions()
| subroutine, public datamodule::load_output_regions | ( | character (len=*), intent(in) | dir, |
| integer, intent(out) | nreg, | ||
| integer, intent(out) | status ) |
Load file with information on output regions; Outregions.txt.
Consequences Module worldvar variable outregion is allocated and set, and variable noutreg is set.
- Parameters
-
[in] dir File directory [out] nreg Number of output regions [out] status Error status
If not allocated: allocate and initialize outregion
◆ load_parameters()
| subroutine, public datamodule::load_parameters | ( | character(len=*), intent(in) | dir, |
| type(basemodeltype), intent(in) | basemodel, | ||
| integer, intent(out) | status, | ||
| logical, intent(in), optional | seqflag, | ||
| integer, intent(in), optional | iens ) |
Read parameter values from file par.txt.
Consequences The modvar module variables soilpar, landpar, genpar, basinpar, regpar, lakedatapar and monthpar will be (re)set.
- Parameters
-
[in] dir File directory [in] basemodel Information on basemodel [out] status Error status of subroutine [in] seqflag Flag for using sequence number in par-file name (parseq) [in] iens Index of ensemble simulation, 0 for ordinary par.txt
Algorithm
Get filename and open par-file
Allocate and initiate parameter variables
Read parameter values from file
Handle submodel for basin parameters
◆ load_permanent_soilleakage()
| subroutine, public datamodule::load_permanent_soilleakage | ( | character (len=*), intent(in) | dir, |
| integer, intent(in) | n, | ||
| type(datetype), intent(in) | bd, | ||
| integer, intent(out) | status ) |
Gets the information about nutrient soil leakage from file.
Alternative 1: Soil nutrient leakage concentration is read from file and will replace the concentration calculated by HYPE soil routines. The runoff from classes gets the new concentration.
Alternative 2: Soil nutrient leakage load is read from file. A simplified soil routine is used.
Consequences Module modvar variable soilleak is allocated and set.
Reference ModelDescription Chapter 4 (Nutrient soil leakage from outer source)
- Parameters
-
[in] dir file directory [in] n number of subbasins [in] bd simulation begin date [out] status error status
Algorithm
Load monthly soilleakage concentrations for subbasins (SMED-HYPE)
- Allocate and initiate variables
- Read data files
Load monthly soilleakage loads per class
- Allocate and initiate variables
- Read data files
Load first year of monthly soilleakage loads per class
- Allocate and initiate variables
- Read data files
Load constant soilleakage loads or concentrations, depending on classmodel (5 or 6)
- Allocate and initiate variables
- Read data files
Load constant or monthly loads or concentrations, depending on classmodel (5 or 6 or 10 or 9)
- Allocate and initiate variables
- Read data files
Monthly time series of loads or concentrations, depending on classmodel (5 or 6 or 10 or 9)
- Allocate and initiate variables
- Read data files
◆ load_pointsourcedata()
| subroutine, public datamodule::load_pointsourcedata | ( | character (len=*), intent(in) | dir, |
| character (len=*), intent(in) | infile, | ||
| integer, intent(in) | n, | ||
| integer, intent(out) | status ) |
Gets the information about nutrient point sources from PointSourceData.txt Recalculate into form suitable for model. Set permanent loads.
Consequences Module modvar variable psinfo or load is set. Module worldvar variable psdates may be allocated and set.
Reference ModelDescription Chapter 5 (Point sources)
- Parameters
-
[in] dir File directory (modeldir) [in] infile Name of pointsource file to be read (PointSourceData.txt) [in] n Number of subbasins [out] status Error status
If periodically constant pointsources are used
- Allocate and initiate variables
- Collect dates where point source load changes; fromdata and todate+1
- Calculate permanent point source loads
- Deallocate load variable for subbasins without point sources
If timeseries of pointsources is used:
- Check if PointSourceTimeSerie.txt exist as assumed from read_pointsourcedata.
- Allocate find corresponding index for subid
- Calculate the number of used positive and negative pointsources, and allocate variable to hold pointsource information
- Set pointsource information variable
Check if timeseries are correct and present, and calculate their index coupling
- Check time periods of pointsource time series
- Prepare PointSourceTimeSeries.txt to be read.
- Prepare PSMonthlySeries.txt to be read.
- Prepare PSYearlySeries.txt to be read.
◆ load_rrcurvedata()
|
private |
Gets the information about parameters to use for calculating main river water level from RiverRatingCurveData.txt.
Consequences Module variables will be allocated and set
- Parameters
-
[in] funit Unit for file [in] dir File directory [in] n Number of subbasins [out] status error status
Algorithm
Check for file and if no file do turn off calc of corl
◆ load_submodel_information()
| subroutine, public datamodule::load_submodel_information | ( | character(len=*), intent(in) | dir, |
| logical, intent(in) | submodelrun, | ||
| type(basemodeltype), intent(inout) | basemodel, | ||
| integer, intent(out) | msub, | ||
| integer, intent(out) | status ) |
Reads file with information about submodel for the current.
- Parameters
-
[in] dir File directory [in] submodelrun Submodel is supposed to be run [in,out] basemodel Information on basemodel [out] msub Number of subbasins of submodel [out] status Error status
◆ prepare_for_update()
| subroutine, public datamodule::prepare_for_update | ( | character(len=*), intent(in) | dir, |
| logical, dimension(dim_update), intent(in) | locupall, | ||
| logical, dimension(dim_update), intent(in) | locupnone, | ||
| integer, intent(in) | nsfile, | ||
| integer, intent(in) | ns, | ||
| integer, intent(out) | status ) |
Read information for basins to update.
Consequences Module modvar variables conduct, doupdate, useinupdate may change.
- Parameters
-
[in] dir File directory [in] locupall Flag for update on all stations [in] locupnone Flag for no update on any stations [in] nsfile Number of subbasins of basemodel [in] ns Number of subbasins [out] status Error number
Algorithm
Read update-file if available
Prepare discharge data stations to use for replacing flow (quseobs)
Prepare dicharge data stations and AR-coefficient to use for AR-updating flow (qar)
Prepare waterstage data stations and AR-coefficient to use for AR-updating flow (war)
Prepare for correction of phosphorus concentration by a factor (tpcorr,tploccorr)
Prepare for correction of nitrogen concentration by a factor (tncorr,tnloccorr)
Prepare concentration data stations to use for replacing simulated concentrations (cuseobs)
Prepare for updating lake waterstage at end of time step (wendupd)
Check for incompatible update methods (war not to be used with qar or wendupd)
Write information about used updating methods to logg-file
◆ prepare_subbasin_output()
| subroutine, public datamodule::prepare_subbasin_output | ( | integer, dimension(:), intent(inout), allocatable | critbasins, |
| integer, intent(out) | status ) |
Preparation for subbasin and regional output files Preparation of outputs regarding subbasins and regions. Selected subbasins/regions for subbasin and regional output files and criteria calculations.
- Parameters
-
[in,out] critbasins Subid of subbasins selected for criteria calculations [out] status Error status
Find and set index of areas to be written as subbasin- or region-files (areas was given as ids) and reduce them to only those in (sub-)model
Find and set index of subbasins to be excluded from criteria calculation
◆ read_and_calc_basindata()
| subroutine, public datamodule::read_and_calc_basindata | ( | integer, intent(in) | funit, |
| character (len=*), intent(in) | infile, | ||
| integer, intent(in) | nskip, | ||
| integer, intent(in) | n, | ||
| integer, intent(in) | maxcol, | ||
| integer, dimension(n), intent(out) | lakedataid, | ||
| integer, intent(out) | status ) |
Gets the information about subbasins from GeoData. Reads the matrix of basin data values from the file vith mcols columns.
Consequences Module variables will be allocated and set
- Parameters
-
[in] funit Unit for file [in] infile Name of characteristics file to be read [in] nskip Number of comment rows [in] n Number of subbasins [in] maxcol Maximum number of data columns [out] lakedataid lakedataid from GeoData [out] status Error status
◆ read_diffuse_sources_file()
|
private |
Gets the information about diffuse sources for subbasins from file like NonPointSourceData.txt Reads the matrix of basin data values from a file with mcols columns Handles missing subbasins (no diffuse load), and extra subid (ignored).
Consequences Module variables will be allocated and set
- Parameters
-
[in] funit Unit for file [in] infile Filename and directory [in] nsbase Number of subbasins in GeoData [in] n Number of subbasins in (sub)model [in] nskip Number of comment rows in file [out] status Error status
◆ read_first_monthly_leakage()
|
private |
Reads monthly time series of soil leakage, first year.
Reference ModelDescription Chapter Nitrogen and phosphorus in land routines (Nutrient soil leakage from outer source)
- Parameters
-
[in] dir File directory [in] infile Name of leakage file [in] bd Simulation begin date [in] n Number of subbasins (base) [in] geoid Subid of subbasins (base) [out] status Error status [in,out] leakage Soil leakage concentrations
◆ read_one_year_monthly_leakage()
|
private |
Reads monthly time series of soil leakage, one year.
Reduce the data for submodel.
Reference ModelDescription Chapter Nitrogen and phosphorus in land routines (Nutrient soil leakage from outer source)
- Parameters
-
[in] dir file directory [in] infile name of leakage file [in] cd current date [in] n number of subbasins [in] geoid subid of subbasins [out] status error status [in,out] leakage soil leakage concentrations
◆ read_parameter_ensemble_task()
|
private |
Reads file with intructions on parameter ensemble simulation.
- Parameters
-
[in] dir File directory [out] taskas task parameter ensemble with allsim.txt [out] taskbs task parameter ensemble with bestsims.txt
Algorithm
Default values
Check for file
Open and read simulation settings from optpar.txt
◆ read_pointsourcedata()
|
private |
Reads the matrix of point source data from the file.
Reference ModelDescription Chapter 5 (Point sources)
- Parameters
-
[in] dir File directory [in] infile Name of pointsource file to be read (PointSourceData.txt) [out] nrows Number of rows in file [out] status Error status [out] fexist Status of file exist [in,out] readsubid subid if point sources [in,out] readpsid pointsource id [in,out] readpstype type of point source [in,out] readlocation location of abstraction/tracer outlet [in,out] pointsource Read pointsource info (subbasins,column) [in,out] readdate1 From date of point source [in,out] readdate2 To date of point source
◆ read_seasonal_leakage()
|
private |
Reads the matrix of soil leakage concentrations or loads from file. The file can have seasonality as daily or monthly values or be constant.
Reads the typical soil leakage and checks them (>=0).
Reference ModelDescription Chapter Nitrogen and phosphorus in land routines (Nutrient soil leakage from outer source)
- Parameters
-
[in] dir File directory [in] infile Name of leakage file [in] n Number of subbasins (basemodel) [in] geoid Subid of subbasins [in] nleak Number of values to read for each subbasin (1, 12 (or 366)) [out] status Error status [in,out] leakage Soil leakage concentrations
◆ read_simulation_perf_and_par()
|
private |
Read the performance result and used parameter values from the one simulation (one row) from a file.
- Parameters
-
[in] funit File unit [in] i Simulation number [in] mpar Dimension of parameters [in] numpar Number of optimised parameters [in] nperf Number of performance measures [in] numcrit Number of optimised variables [in,out] optcrit Value of optimation criterion for the simulation [in,out] performance performance criteria for the simulation [out] parameters Parameter values for the simulation [out] status flag for subroutine status
◆ read_update_data()
|
private |
Gets the information about update from update.txt.
- Parameters
-
[in] funit Unit for file [in] infile Name of characteristics file to be read [in] ns Number of subbasins (basemodel) [out] nqsub Number of subbasins with quseobs update [out] qupdsub Subid of subbasins with quseobs update [out] nqarsub Number of subbasins with qAR date [out] nwarsub Number of subbasins with wAR date [out] qarupdsub Subid of subbasins with qAR update [out] warupdsub Subid of subbasins with wAR update [out] qarupdar AR-factor for qAR-update [out] warupdar AR-factor for wAR-update [out] nwsub Number of subbasins with wendupd update [out] wendsub Subid of subbasins with wendupd update [out] nrow Number of data rows in file [out] tpcorrarr Subid,value of subbasins with tpcorr update [out] tncorrarr Subid,value of subbasins with tncorr update [out] tploccorrarr Subid,value of subbasins with tploccorr update [out] tnloccorrarr Subid,value of subbasins with tnloccorr update [out] ncsub Number of subbasins with cuseobs update [out] cupdsub Subid of subbasins with cuseobs update
◆ reform_inputdata_for_submodel()
| subroutine, public datamodule::reform_inputdata_for_submodel | ( | type(basemodeltype), intent(in) | basemodel, |
| integer, intent(in) | ns ) |
Reform the information about subbasins to arrays for submodel.
- Parameters
-
[in] basemodel Information on basemodel [in] ns number of subbasins to be simulated
◆ reload_classdata()
| subroutine, public datamodule::reload_classdata | ( | integer, intent(in) | funit, |
| character (len=*), intent(in) | dir, | ||
| type(datetype), intent(in), optional | filedate, | ||
| integer, intent(out) | status, | ||
| logical, dimension(2), intent(out) | statuscdc ) |
Gets the information about classes from ClassData_yyyymmdd.txt.
Consequences Module variable classdata will be reset
- Parameters
-
[in] funit Unit for file [in] dir File directory [in] filedate date of start of this file [out] status File found and read ok [out] statuscdc Changed classes and soiltypes found
Algoritm
Check for file.
Allocate local variables for holding class information
Read information of ClassData
- Skip comment lines in the beginning of the file
- Read column headings, and find the column with class number
- Read class information, and sort according to class order.
Collect class information in column sorted array. Only these columns are allowed to change
Replace classdata values for found classes
◆ reload_deposition()
| subroutine, public datamodule::reload_deposition | ( | integer, intent(in) | funit, |
| character (len=*), intent(in) | dir, | ||
| type(basemodeltype), intent(inout) | basemodel, | ||
| integer, intent(in) | n, | ||
| integer, intent(out) | status, | ||
| type(datetype), intent(in), optional | filedate ) |
Gets the information about atmospheric deposition from AtmdepData.
Consequences Module variables deposition2 may be reallocated and reset
- Parameters
-
[in] funit Unit for file [in] dir File directory [in,out] basemodel Information on basemodel [in] n Number of subbasins (sub)model [out] status error status [in] filedate date of start of this file
Check for substance simulation
Check for file and return if no file
◆ reload_diffuse_sources()
| subroutine, public datamodule::reload_diffuse_sources | ( | integer, intent(in) | funit, |
| character (len=*), intent(in) | dir, | ||
| integer, intent(in) | nsbase, | ||
| integer, intent(in) | n, | ||
| integer, intent(out) | status, | ||
| type(datetype), intent(in) | filedate ) |
Gets the information about diffuse sources for subbasins from NonPointSourceData file of a specified beginning date. These are used until next file is read.
- Parameters
-
[in] funit Unit for file [in] dir File directory [in] nsbase Number of subbasins in GeoData [in] n Number of subbasins in (sub)model [out] status Error status [in] filedate date of start of this file
Check for file and return if no file
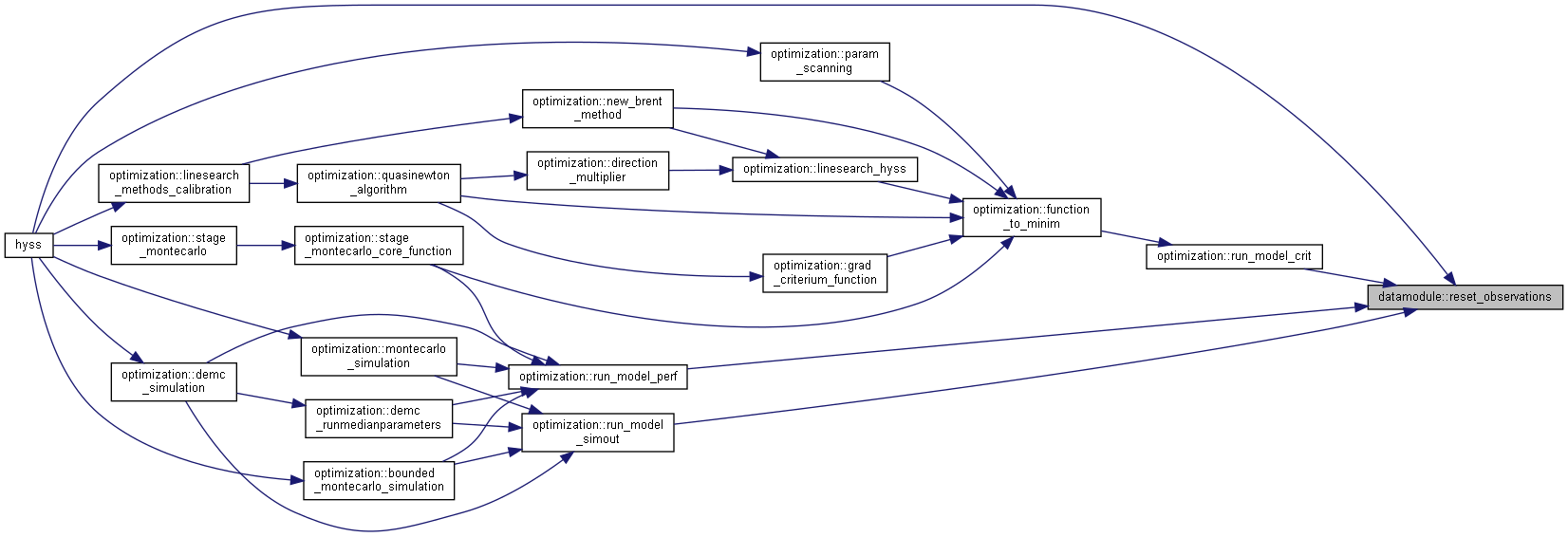
◆ reset_observations()
| subroutine, public datamodule::reset_observations | ( | character(len=*), intent(in) | dir, |
| character(len=*), intent(in) | mdir, | ||
| character(len=*), intent(in) | odir, | ||
| integer, intent(out) | status ) |
Prepare forcingdata, point source time series and other observations for another simulation in case they are read each time step.
- Parameters
-
[in] dir File directory (forcingdir) [in] mdir File directory (modeldir) [in] odir File directory (otherobsdir) [out] status Error number
Algorithm
Reset observation files
Reset model defined observations
Reset point sources time files
◆ save_xoreginformation()
|
private |
Save information about finding other observation series.
Consequences Module index array (xoregindex) is set.
- Parameters
-
[in] n Number of columns in Xoregobs [in] var2 Variables and subid for Xoregobs columns [in] nr Number of outregions
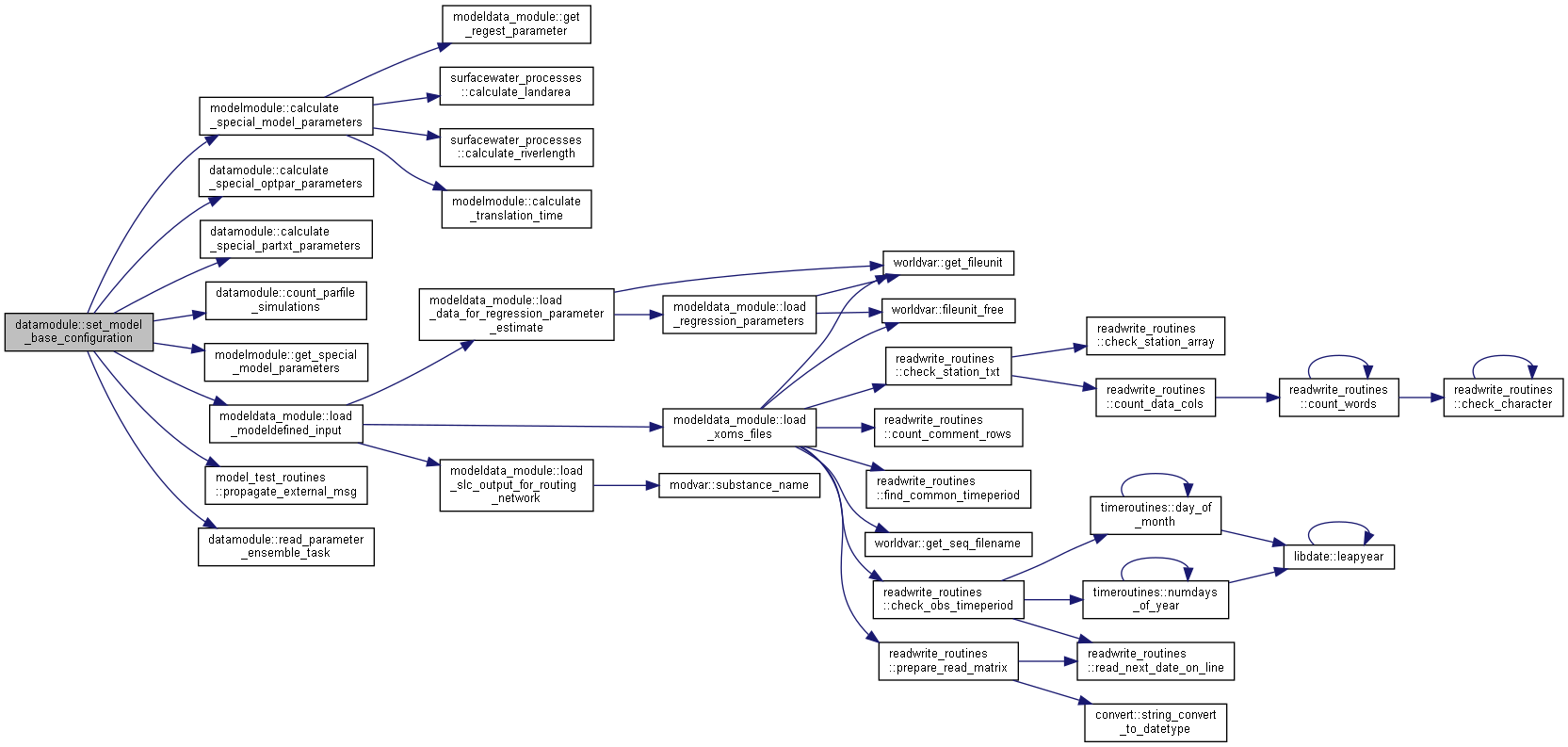
◆ set_model_base_configuration()
| subroutine, public datamodule::set_model_base_configuration | ( | type(basemodeltype), intent(in) | basemodel, |
| character(len=*), intent(in) | idir, | ||
| character(len=*), intent(in) | idir2, | ||
| type(statedimensiontype), intent(inout) | dim, | ||
| integer, intent(out) | status ) |
Set model base configuration.
Handle dimension of river queue state; the queue length may be different for current model, initial state file, submodel and for optimization. Set structure holding state dimensions. Transform crop information for southern hemisphere.
Consequences Module modvar variables cropdata and cropirrdata may be changed.
- Parameters
-
[in] basemodel Information on basemodel [in] idir file directory (modeldir) [in] idir2 file directory (otherobsdir) [in,out] dim simulation dimensions [out] status error status of subroutine
Algorithm
Initialisations
Find maximum dimension of river queue variables
Set state dimension variable
Check maximum number of lakesections to set for connectivity fill-and-spill ilake or HDS model
Set current substances minimum and maximum concentration limits
Check if river queue variables is compatible with initial state and calibration
◆ set_model_configuration()
| subroutine, public datamodule::set_model_configuration | ( | type(stateconfigurationtype), intent(inout) | config, |
| type(simulationconfigurationtype), intent(inout) | simconfig ) |
Set model choice configuration and state dimensions.
- Parameters
-
[in,out] config simulation configuration [in,out] simconfig simulation configuration
◆ set_outvar()
| subroutine, public datamodule::set_outvar | ( | integer, intent(in) | nout, |
| integer, intent(out) | noutvar, | ||
| integer, intent(out) | noutvarclass ) |
Saves information on variables to calculate for output.
- Parameters
-
[in] nout number of output file type to be set [out] noutvar number of output variables (so far) [out] noutvarclass number of output variables for class output (so far)
◆ set_outvar_crit()
|
private |
Saves information on output variables to calculate for use in criteria calculations.
Consequences Module variable acccalvar, outvarinfotemp, noutvar and outvarindex is changed.
- Parameters
-
[in,out] noutvar number of output variables to be set (so far) [out] noutvarclass number of output variables for class output (so far)
Algorithm
◆ set_outvar_for_variable_new()
| subroutine, public datamodule::set_outvar_for_variable_new | ( | integer, intent(in) | varid, |
| integer, intent(in) | areaagg, | ||
| integer, intent(out) | ovindex, | ||
| integer, intent(inout) | noutvar, | ||
| integer, intent(inout), optional | noutvarclass, | ||
| character(len=6), intent(in), optional | cgname ) |
Turns on calculation of choosen variable.
- Parameters
-
[in] varid index in outvarid of current variable [in] areaagg aggregation of variable (0=subbasin average, 1=upstream, 2=region) [out] ovindex index of current variable in outvar [in,out] noutvar number of output variables to be set (so far) [in,out] noutvarclass number of class output variables to be calculated (so far) [in] cgname class group name
Algorithm
Increase the counter of number of output variables to calculate
◆ set_outvar_output()
|
private |
Saves information on variables to write to output files.
Consequences Module variable outvarinfotemp and outvarindex is allocated.
- Parameters
-
[in] nout number of output file type to be set [out] noutvar number of output variables (so far) [out] noutvarclass number of output variables for class output (so far)
Algorithm
Count number of classoutputs
Initialize and allocate variables holding outvar information
For every output file type...
and every variable..
- Set outvar information
◆ set_outvar_test_information()
| subroutine, public datamodule::set_outvar_test_information | ( | integer, intent(in) | n | ) |
Add information of test of output.
- Parameters
-
[in] n number of output variables
◆ set_update_nutrientcorr()
|
private |
Set variable for phosphorus and nitrogen correction/updating.
Consequences Module variables doupdate and useinupdate are allocated and set.
- Parameters
-
[in] dim number of subbasins of base model [in] ns number of subbasins [in] nrow number of data [in] icorr index for type of updating/correction [in] dataarray read data
Algorithm
For each subbasin and data point: If equal set correction value for this subbasin
If no correction value are found: turn off updating and deallocate array
◆ set_yesno_variable_from_line()
|
private |
Read Y/J/N code from line to set logical variable.
- Parameters
-
[in] linelen length of line [in,out] linepos current position on line [in] line line to be read [in] errstr value of str if error [out] onoff value of yesno variable [out] status error statur of subroutine
◆ start_lakedata_table()
|
private |
Set lakedatapar values to those model variable values specified in LakeData.txt.
Consequences Module modvar variables lakedatapar_ini,lakedatapar, lakedataparindex may be allocated and set.
- Parameters
-
[in] maxcol Maximum possible amount of columns on LakeData.txt [in] ncols Number of columns effectively used in LakeData.txt [in] maxrow Maximum number of rows in LakeData.txt (nsub) [in] nrows Number of rows effectively used in LakeData.txt [in] str Character array with labels of ALL columns given in LakeData.txt (column headers) [in] xi Integer values aquired by reading LakeData.txt [in] xr Float values aquired by reading LakeData.txt [in] iindex Index for column real variables [in] rindex Index for column integer variables [in] ns Number of subbasins (base model) [in] lakedataid lakedataid from GeoData.txt
◆ turn_off_soilleakage()
|
private |
Check if substances using load or leak files are simulated.
Algorithm
Turn off soilleakage if no substances simulated
Turn off soilleakage if only T2 is simulated
Set output value
Generated by