Functions/Subroutines | |
| subroutine, public | prepare_outputfiles (dir, n, na, iens, runens, allsim, ensstat) |
| subroutine, public | close_outputfiles (n, na, iens, ensstat) |
| subroutine | open_subbasinfiles (dir, n, iens, runens, allens, ensstat) |
| subroutine | create_filename_for_basin (filename, basinid, usesuffix1, suffix1, suffix2, suffix2format, suffix3, suffix4) |
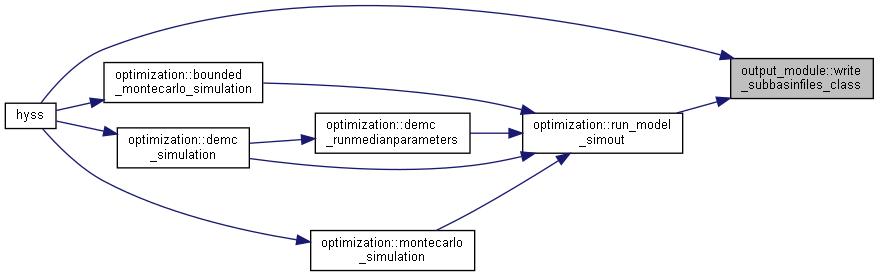
| subroutine, public | write_subbasinfiles (io, idt, ndt, ct) |
| subroutine, public | write_subbasinfiles_in_parallel (io, idt, ndt, iens, ct) |
| subroutine, public | write_regionfiles (io, idt, ndt, ct) |
| subroutine, public | write_regionfiles_in_parallel (io, idt, ndt, iens, ct) |
| subroutine, public | write_subbasinfiles_class (io, idt, ndt, ct) |
| subroutine, public | write_subbasinfiles_class_in_parallel (io, idt, ndt, iens, ct) |
| subroutine | close_subbasinfiles (n, iens, ensstat) |
| subroutine | open_timefiles (dir, n, iens, runens, allens, ensstat) |
| subroutine | open_timefiles_classes (dir, n, iens, runens, allens, ensstat) |
| subroutine, public | write_timefiles (io, idt, ndt, ct) |
| subroutine, public | write_timefiles_netcdf (io, idt, ndt, ct) |
| subroutine, public | write_timefiles_in_parallel (io, idt, ndt, iens, ct) |
| subroutine, public | write_timefiles_netcdf_in_parallel (io, idt, ndt, iens, ct) |
| subroutine | close_timefiles (iens, ensstat) |
| subroutine | close_timefiles_class (iens, ensstat) |
| subroutine, public | save_mapfiles (dir, ntime, iens, runens, allens) |
| subroutine, public | save_loadfiles (dir, year) |
| subroutine, public | write_subbasin_assessment (filedir, na, sas2, iens, runens) |
| subroutine, public | write_simulation_assessment (dir, iens, n, optcrit, performance, runens, ccrit, cthres) |
| subroutine, public | save_respar (dir, numpar, n) |
| subroutine | get_parametervalues (varindex, n, dim, values, num) |
| subroutine, public | prepare_save_all_simulations (dir, filename, funit, numpar, nperf, numcrit) |
| subroutine | write_ensemble_simulations_heading (funit, numpar, nperf, numcrit, popflag) |
| subroutine, public | save_ensemble_simulations (dir, numpar, nperf, numcrit, nsim, optcrit, performance, parameters) |
| subroutine, public | write_simulation_results_perf_and_par (funit, i, mpar, numpar, nperf, numcrit, optcrit, performance, parameters, mlab, jpop, igen, iacc) |
Detailed Description
Prepare for and save result.
Procedures for preparing output data and for saving result files
Function/Subroutine Documentation
◆ close_outputfiles()
| subroutine, public output_module::close_outputfiles | ( | integer, intent(in) | n, |
| integer, intent(in) | na, | ||
| integer, intent(in) | iens, | ||
| integer, intent(in), optional | ensstat ) |
Manage standard output files; close files.
- Parameters
-
[in] n Number of subbasins [in] na Number of aquifers [in] iens Current simulation [in] ensstat Flag for writing parallell ensemble files
◆ close_subbasinfiles()
|
private |
Close files for subbasin and output region printout.
- Parameters
-
[in] n Number of subbasins [in] iens Current simulation [in] ensstat Flag for writing parallell ensemble files
◆ close_timefiles()
|
private |
Close files for timeserie output.
- Parameters
-
[in] iens Current simulation [in] ensstat Flag for writing parallell ensemble files
◆ close_timefiles_class()
|
private |
Close files for timeserie output for classes.
- Parameters
-
[in] iens Current simulation [in] ensstat Flag for writing parallell ensemble files
◆ create_filename_for_basin()
|
private |
Create file name string for subbasin- or outregion file.
- Parameters
-
[out] filename file name [in] basinid subid or outregid; to be name of file [in] usesuffix1 status of using suffix1 [in] suffix1 string file suffix [in] suffix2 integer file suffix [in] suffix2format use 6 character suffix instead of three [in] suffix3 integer suffix for classoutput [in] suffix4 classgroup name suffix
◆ get_parametervalues()
|
private |
Collect parameter values from model variables.
- Parameters
-
[in] varindex model parameter index [in] n number of subbasins [in] dim max number of parameter values (soil types/land uses/subbasins) [out] values parameter values [out] num number of parameter values (soil types/land uses/subbasins) that are used
Algoritm
Depending on variable type: Find variable dimension and parameter values
◆ open_subbasinfiles()
|
private |
Opens files for subbasin and output region printout, including classoutput.
- Parameters
-
[in] dir Result file directory [in] n Number of subbasins [in] iens Current simulation [in] runens Flag for ensemble simulation [in] allens Flag for writing all ensemble results, i.e. 6 characters for integer [in] ensstat Flag for writing parallell ensemble files
◆ open_timefiles()
|
private |
Opens files for timeserie printout.
- Parameters
-
[in] dir File directory [in] n Number of data columns (subbasins) [in] iens Current simulation [in] runens Flag for ensemble run [in] allens Flag for writing all ensemble results [in] ensstat Flag for writing parallell ensemble files
◆ open_timefiles_classes()
|
private |
Opens files for timeserie printout of classes.
- Parameters
-
[in] dir File directory [in] n Number of data columns (subbasins) [in] iens Current simulation [in] runens Flag for ensemble run [in] allens Flag for writing all ensemble results [in] ensstat Flag for writing parallell ensemble files
◆ prepare_outputfiles()
| subroutine, public output_module::prepare_outputfiles | ( | character(len=*), intent(in) | dir, |
| integer, intent(in) | n, | ||
| integer, intent(in) | na, | ||
| integer, intent(in) | iens, | ||
| logical, intent(in) | runens, | ||
| logical, intent(in) | allsim, | ||
| integer, intent(in), optional | ensstat ) |
Manage standard output files; opens files and write heading.
- Parameters
-
[in] dir Result file directory [in] n Number of subbasins [in] na Number of aquifers [in] iens Current simulation [in] runens Flag for using ensemble number of ensemble simulation [in] allsim Flag for writing all simulation results [in] ensstat Flag for writing parallell ensemble files
◆ prepare_save_all_simulations()
| subroutine, public output_module::prepare_save_all_simulations | ( | character(len=*), intent(in) | dir, |
| character(len=*), intent(in) | filename, | ||
| integer, intent(in) | funit, | ||
| integer, intent(in) | numpar, | ||
| integer, intent(in) | nperf, | ||
| integer, intent(in) | numcrit ) |
Prepare a file to writes result from all MonteCarlo simulations.
- Parameters
-
[in] dir File directory [in] funit Unit to be connected to file [in] numpar Number of optimised parameters [in] nperf Number of performance measures [in] numcrit Number of optimised variables
◆ save_ensemble_simulations()
| subroutine, public output_module::save_ensemble_simulations | ( | character(len=*), intent(in) | dir, |
| integer, intent(in) | numpar, | ||
| integer, intent(in) | nperf, | ||
| integer, intent(in) | numcrit, | ||
| integer, intent(in) | nsim, | ||
| real, dimension(nsim), intent(in) | optcrit, | ||
| real, dimension(nsim,nperf,numcrit), intent(in) | performance, | ||
| real, dimension(nsim,numpar), intent(in) | parameters ) |
Write to file result from the nsim best MonteCarlo simulations.
- Parameters
-
[in] dir File directory [in] numpar Number of optimised parameters [in] nperf Number of performance measures [in] numcrit Number of optimised variables [in] nsim Number of simulations to be saved [in] optcrit Optimation criterion for simulations [in] performance Performance measures for simulations [in] parameters Parameter values for simulations
◆ save_loadfiles()
| subroutine, public output_module::save_loadfiles | ( | character(len=*), intent(in) | dir, |
| integer, intent(in) | year ) |
Save the loads for the last year to files. Subroutine is called once per year. Could be the load of a part (period) of the last year.
Consequences Module worldvar variable accload are zeroed.
- Parameters
-
[in] dir File directory [in] year Current year
Algorithm
Preparation to write heading
Save data to one file per substance (not including T2)
- Open file
- Write heading prepared above
- Write data for each subbasin on rows
Reset accumulation variables to zero at end of year (or output period)
◆ save_mapfiles()
| subroutine, public output_module::save_mapfiles | ( | character(len=*), intent(in) | dir, |
| integer, intent(in) | ntime, | ||
| integer, intent(in) | iens, | ||
| logical, intent(in) | runens, | ||
| logical, intent(in) | allens ) |
Writes the files with data in format suitable for mapping.
- Parameters
-
[in] dir File directory [in] ntime Number of values in vector = number of timesteps [in] iens Current simulation [in] runens Flag for ensemble simulation [in] allens Flag for writing all ensemble results
◆ save_respar()
| subroutine, public output_module::save_respar | ( | character(len=*), intent(in) | dir, |
| integer, intent(in) | numpar, | ||
| integer, intent(in) | n ) |
Save optimal values of optimized parameters to file respar.txt.
- Parameters
-
[in] dir File directory [in] numpar number of parameters that has been calibrated [in] n number of subbasins
Algoritm
Allocate and initiate local variables
Open file for writing parameter and write heading
For every calibrated parameter:
- Get current parameter value
- Write parameter to file
◆ write_ensemble_simulations_heading()
|
private |
Write heading to file for MonteCarlo simulations.
- Parameters
-
[in] funit File unit [in] numpar Number of optimised parameters [in] nperf Number of performance measures [in] numcrit Number of optimised variables [in] popflag Flag for population columns included
◆ write_regionfiles()
| subroutine, public output_module::write_regionfiles | ( | integer, intent(in) | io, |
| integer, intent(in) | idt, | ||
| integer, intent(in) | ndt, | ||
| type(timeinformationtype), intent(in) | ct ) |
Calculate and write output region files.
- Parameters
-
[in] io Current output [in] idt Current time step [in] ndt Maximum simulation time steps [in] ct Current time
◆ write_regionfiles_in_parallel()
| subroutine, public output_module::write_regionfiles_in_parallel | ( | integer, intent(in) | io, |
| integer, intent(in) | idt, | ||
| integer, intent(in) | ndt, | ||
| integer, intent(in) | iens, | ||
| type(timeinformationtype), intent(in) | ct ) |
Calculate and write output region files.
- Parameters
-
[in] io Current output [in] idt Current time step [in] ndt Maximum simulation time steps [in] iens Current simulation [in] ct Current date-time
◆ write_simulation_assessment()
| subroutine, public output_module::write_simulation_assessment | ( | character(len=*), intent(in) | dir, |
| integer, intent(in) | iens, | ||
| integer, intent(in) | n, | ||
| real, intent(in) | optcrit, | ||
| real, dimension(maxperf,n), intent(in) | performance, | ||
| logical, intent(in) | runens, | ||
| real, intent(in) | ccrit, | ||
| real, intent(in) | cthres ) |
Writes the simulation assessment to hyss file and separate file.
- Parameters
-
[in] dir File directory [in] iens Current simulation [in] n Dimension of performance measures [in] optcrit Opimization criterion value [in] performance Perfomance criteria [in] runens Flag for ensemble simulation [in] ccrit conditional criteria [in] cthres conditional criteria threshold
◆ write_simulation_results_perf_and_par()
| subroutine, public output_module::write_simulation_results_perf_and_par | ( | integer, intent(in) | funit, |
| integer, intent(in) | i, | ||
| integer, intent(in) | mpar, | ||
| integer, intent(in) | numpar, | ||
| integer, intent(in) | nperf, | ||
| integer, intent(in) | numcrit, | ||
| real, intent(in) | optcrit, | ||
| real, dimension(nperf,numcrit), intent(in) | performance, | ||
| real, dimension(mpar), intent(in) | parameters, | ||
| logical, intent(in) | mlab, | ||
| integer, intent(in), optional | jpop, | ||
| integer, intent(in), optional | igen, | ||
| integer, intent(in), optional | iacc ) |
Write the performance result and used parameter values from the last simulations to a file.
- Parameters
-
[in] funit File unit [in] i Simulation number [in] mpar Dimension of parameters [in] numpar Number of optimised parameters [in] nperf Number of performance measures [in] numcrit Number of optimised variables [in] optcrit Value of optimation criterion for the simulation [in] performance performance criteria for the simulation [in] parameters Parameter values for the simulation [in] mlab MATLAB format for print out [in] jpop Population Index in DE-MC simulation [in] igen Generation Index in DE-MC simulation [in] iacc Acceptance Index in DE-MC simulation
◆ write_subbasin_assessment()
| subroutine, public output_module::write_subbasin_assessment | ( | character(len=*), intent(in) | filedir, |
| integer, intent(in) | na, | ||
| real, dimension(:,:,:), intent(in) | sas2, | ||
| integer, intent(in) | iens, | ||
| logical, intent(in) | runens ) |
Writes subbasin/output region assessment to file subassX.txt for each pair of compared variables.
- Parameters
-
[in] filedir File directory [in] na dimension of performance measures (variables) [in] sas2 assessment values [in] iens Current simulation [in] runens Flag for ensemble simulation
Algorithm
Set number format, default is four decimals
Set subass writing format, number of columns
For every pair of variables to be compared
- Determine the filename and open a file
- Create comment row of file
- Create heading of columns
- Write heading to file
- Write data to file
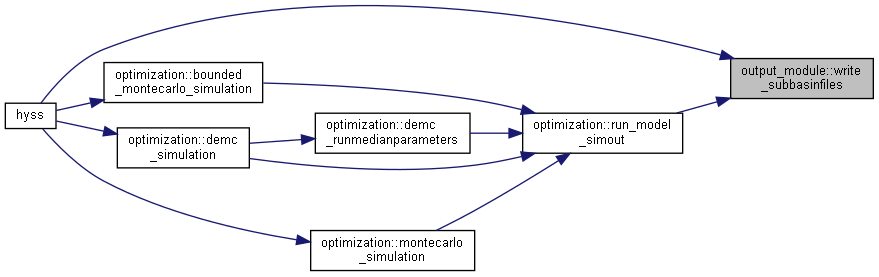
◆ write_subbasinfiles()
| subroutine, public output_module::write_subbasinfiles | ( | integer, intent(in) | io, |
| integer, intent(in) | idt, | ||
| integer, intent(in) | ndt, | ||
| type(timeinformationtype), intent(in) | ct ) |
Calculate and write subbasin files.
- Parameters
-
[in] io Current output [in] idt Current time [in] ndt Maximum simulation time [in] ct current time
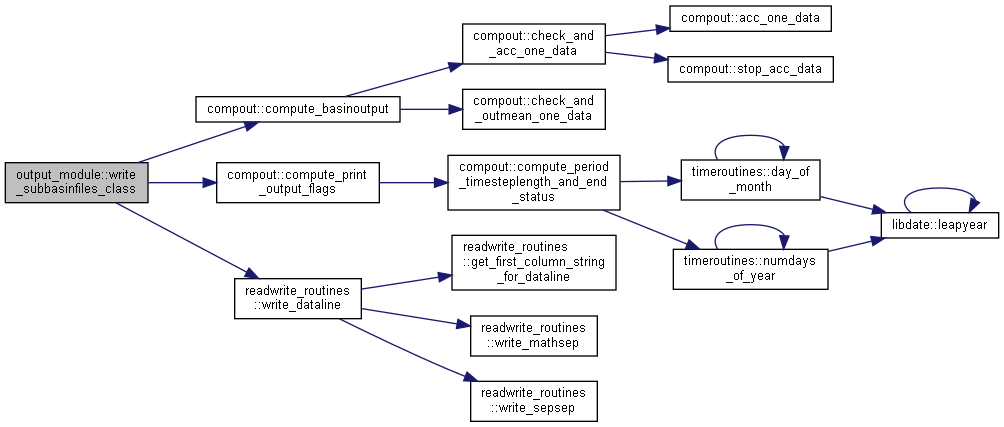
◆ write_subbasinfiles_class()
| subroutine, public output_module::write_subbasinfiles_class | ( | integer, intent(in) | io, |
| integer, intent(in) | idt, | ||
| integer, intent(in) | ndt, | ||
| type(timeinformationtype), intent(in) | ct ) |
Calculate and write subbasin files.
- Parameters
-
[in] io Current output [in] idt Current time [in] ndt Maximum simulation time [in] ct current time
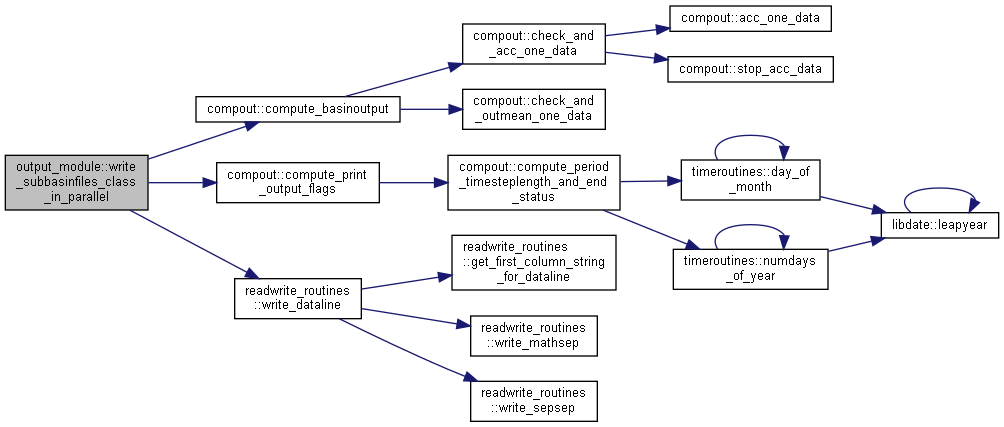
◆ write_subbasinfiles_class_in_parallel()
| subroutine, public output_module::write_subbasinfiles_class_in_parallel | ( | integer, intent(in) | io, |
| integer, intent(in) | idt, | ||
| integer, intent(in) | ndt, | ||
| integer, intent(in) | iens, | ||
| type(timeinformationtype), intent(in) | ct ) |
Calculate and write subbasin files (special for assimilation) Writes sequences' files in parallel.
- Parameters
-
[in] io Current output [in] idt Current time [in] ndt Maximum simulation time [in] iens Current simulation [in] ct current time
◆ write_subbasinfiles_in_parallel()
| subroutine, public output_module::write_subbasinfiles_in_parallel | ( | integer, intent(in) | io, |
| integer, intent(in) | idt, | ||
| integer, intent(in) | ndt, | ||
| integer, intent(in) | iens, | ||
| type(timeinformationtype), intent(in) | ct ) |
Calculate and write subbasin files (special for assimilation) Writes sequences' files in parallel.
- Parameters
-
[in] io Current output [in] idt Current time step [in] ndt Maximum simulation time [in] iens Current simulation [in] ct current time
◆ write_timefiles()
| subroutine, public output_module::write_timefiles | ( | integer, intent(in) | io, |
| integer, intent(in) | idt, | ||
| integer, intent(in) | ndt, | ||
| type(timeinformationtype), intent(in) | ct ) |
Calculate time aggregate of time-output and writes to file.
- Parameters
-
[in] io Current output [in] idt Current time [in] ndt Maximum simulation time [in] ct current time
◆ write_timefiles_in_parallel()
| subroutine, public output_module::write_timefiles_in_parallel | ( | integer, intent(in) | io, |
| integer, intent(in) | idt, | ||
| integer, intent(in) | ndt, | ||
| integer, intent(in) | iens, | ||
| type(timeinformationtype), intent(in) | ct ) |
Calculate time aggregate of time-output and writes to file (special for data assimilation).
- Parameters
-
[in] io Current output [in] idt Current timestep [in] ndt Maximum simulation time [in] iens Current simulation [in] ct current time
◆ write_timefiles_netcdf()
| subroutine, public output_module::write_timefiles_netcdf | ( | integer, intent(in) | io, |
| integer, intent(in) | idt, | ||
| integer, intent(in) | ndt, | ||
| type(timeinformationtype), intent(in) | ct ) |
Calculate time aggregate of time-output and writes to file.
- Parameters
-
[in] io Current output [in] idt Current time [in] ndt Maximum simulation time [in] ct current time
◆ write_timefiles_netcdf_in_parallel()
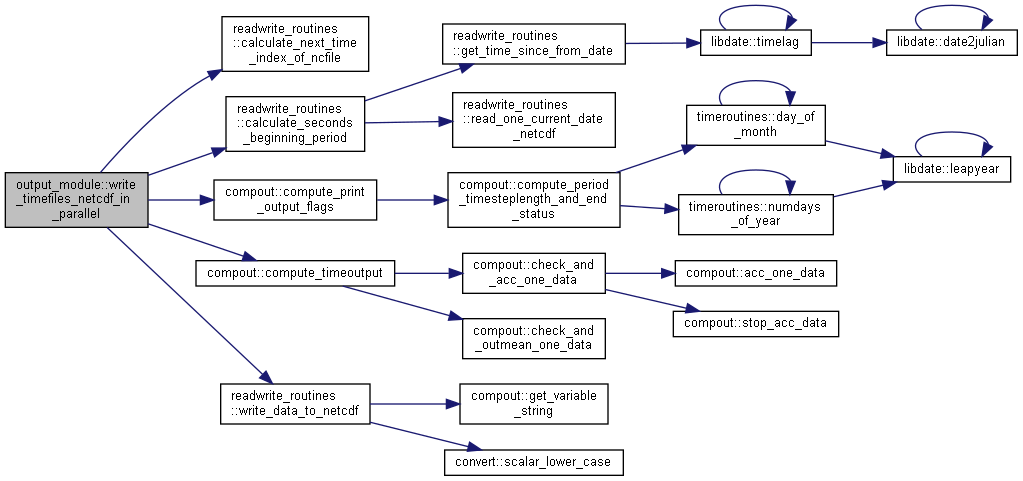
| subroutine, public output_module::write_timefiles_netcdf_in_parallel | ( | integer, intent(in) | io, |
| integer, intent(in) | idt, | ||
| integer, intent(in) | ndt, | ||
| integer, intent(in) | iens, | ||
| type(timeinformationtype), intent(in) | ct ) |
Calculate time aggregate of time-output and writes to file (special for data assimilation).
- Parameters
-
[in] io Current output [in] idt Current time [in] ndt Maximum simulation time [in] iens Current simulation [in] ct current time
Generated by